Get Better Coverage and Discover More Variants with Deep Exome Sequencing
Get Better Coverage and Discover More Variants with Deep Exome Sequencing
Most disease-related variations are located within exons. Exome sequencing captures both common and rare variants missed in traditional GWAS studies. With deep sequencing, researchers can get better coverage (Fig. 1) and discover more variants (Fig. 2); therefore, gaining more insights into the genetic basis of various kinds of cancers and complex diseases. Finding mutations in cancer is particularly challenging because multiple clones may exist within the tumor or only a small percentage of the biopsy may be tumorous. We can overcome these challenges and find those mutations with deep exome sequencing.
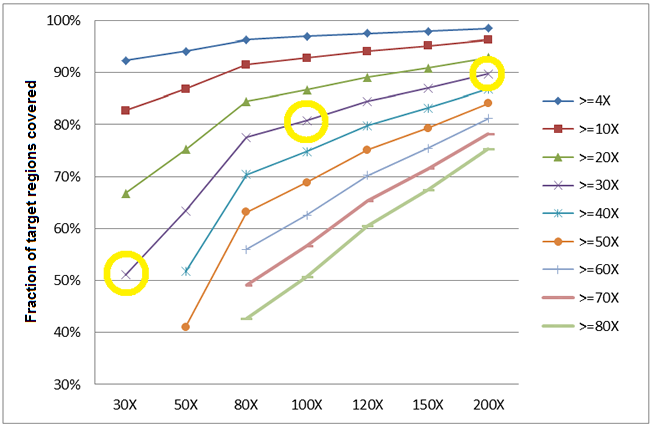
Fig.1 Correlation between the percentage of target regions covered and the sequencing depth in human exome sequencing. Take >=30X series (the purple line) for example: when the sequencing depth is 30X, only half of the target regions (51%) are covered at above 30X. While at the 100X and 200X sequencing depths, a much higher percentage (81% and 90%, respectively) of the target regions is covered at above 30X. The data used in the analysis for each coverage level listed on the X axis are generated by randomly extracting, from a 200X exome sequencing result, the amount of data equivalent to the corresponding coverage level.
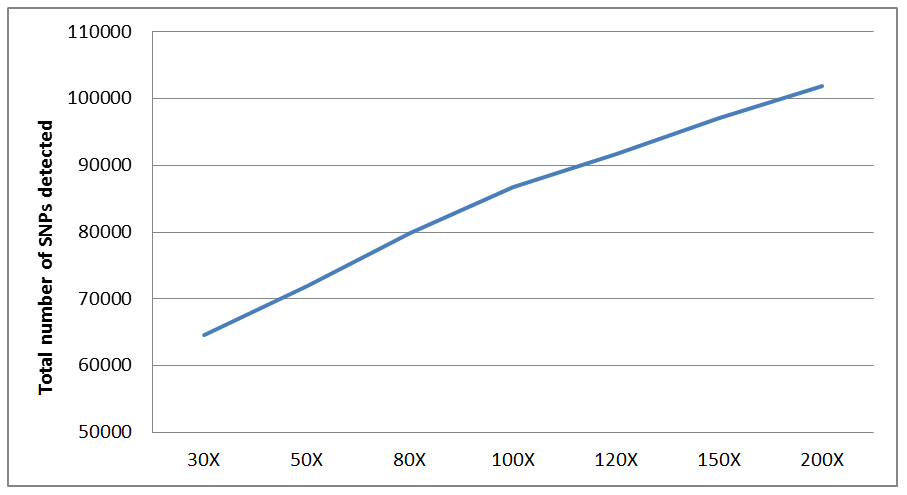
Fig. 2 Correlation between the number of SNPs (detected by the SOAPsnp program) and the sequencing depth. More SNPs are detected at a higher sequencing depth.
Why choose BGI?
- Highly experienced: More than 30,000 human exomes sequenced by BGI to date
- Rapid turnaround at our Hong Kong and CHOP (Philadelphia, PA) facilities
- Affordable pricing
- Strong analytical capabilities: 1,000 bioinformaticians to analyze your data
- High quality data:
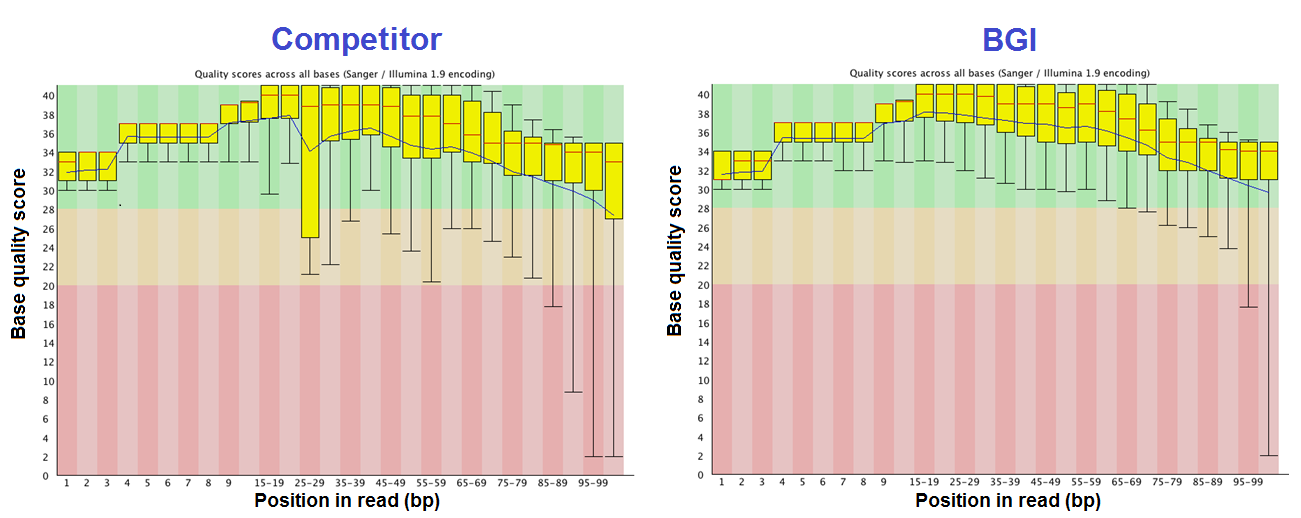
Fig. 3 The Base Quality Plots show that BGI’s data have smaller error bars and higher base quality scores than the competitor’s.
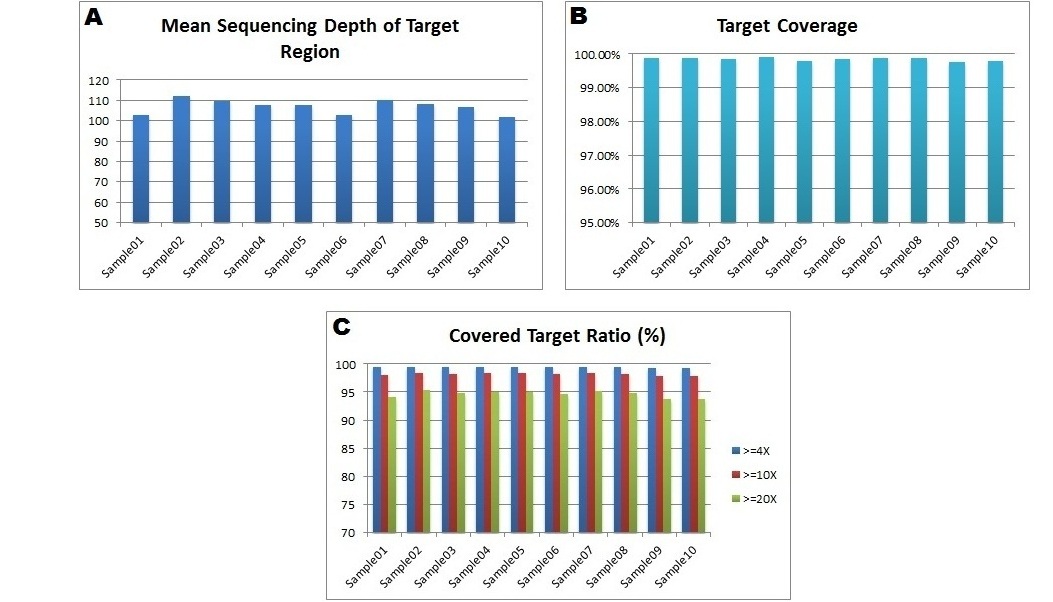
Fig. 1. An example of deep exome sequencing data generated at BGI@CHOP Joint Genome Center using the Agilent SureSelect V4 (51 Mb) Capture Kit. (A) The mean sequencing depth of the targeted region was above 100X for 10 samples. (B) A high coverage (close to 100%) of the 51 Mb target region was achieved across all samples (C) Approximately 95% of the target region had a coverage above 20X, suggesting the high quality and evenness of the data generated.
Case/Publication:
Guo, Guangwu, et al. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nature Genetics. 44,17-19 (2012).