
Cloud Computing
BGI is constantly developing the hardware and software to handle the huge volumes of bioinformatics data using HPC (High Performing Computing) and Cloud computing, and aims to process massive amounts of data with lower cost and higher efficiency to help researchers from all over the world facilitate their research work.
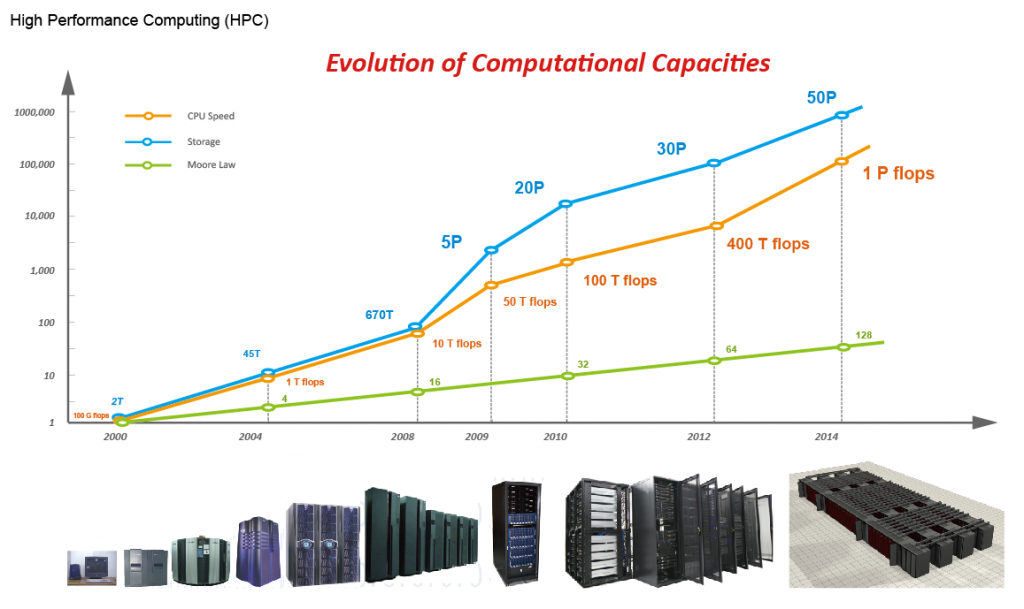 Figure 1. Improvement of computing & storage capability in BGI
Figure 1. Improvement of computing & storage capability in BGI
Two Centers of Cloud Computing in BGI
Currently, there are two large centers in BGI’s cloud computing platform — Bio-Data Centre (CliMB) and Biol-Cloud Computing centre (BGI Cloud).
Bio-data Centre - CliMB
CLiMB is a bio-data center that was independently designed by BGI. The core features include data service, sequence alignment and many others. Building a world-class bio-data center will provide the data support for the health and sustainable development of life science industry and provide high-quality data for researchers from the biology field. CLiMB also supports the data service for Giga Science, a journal published by BGI in partnership with BioMed Central.
Linkage: http://climb.genomics.org.cn
Bio- Cloud Computing Centre - BGI Cloud
BGI Cloud applies advanced technology for cloud storage and cloud computing, integrating the common data and valuable, specific sequencing data of BGI in the field of genomics. BGI also deploys standard bioinformatics analysis pipelines and many kinds of alignment and analysis software to build a set of easy-to-use, flexible, one-stop automatic services for mass data storage and computing. Users are able to remotely conduct the sequencing data processing through the network and get access to the bioinformatics analysis resource anytime anywhere. By integrating powerful computing hardware and rich application software, BGI Cloud provides an end-to-end computing service solution to users around the world.
BGI Cloud Workflow
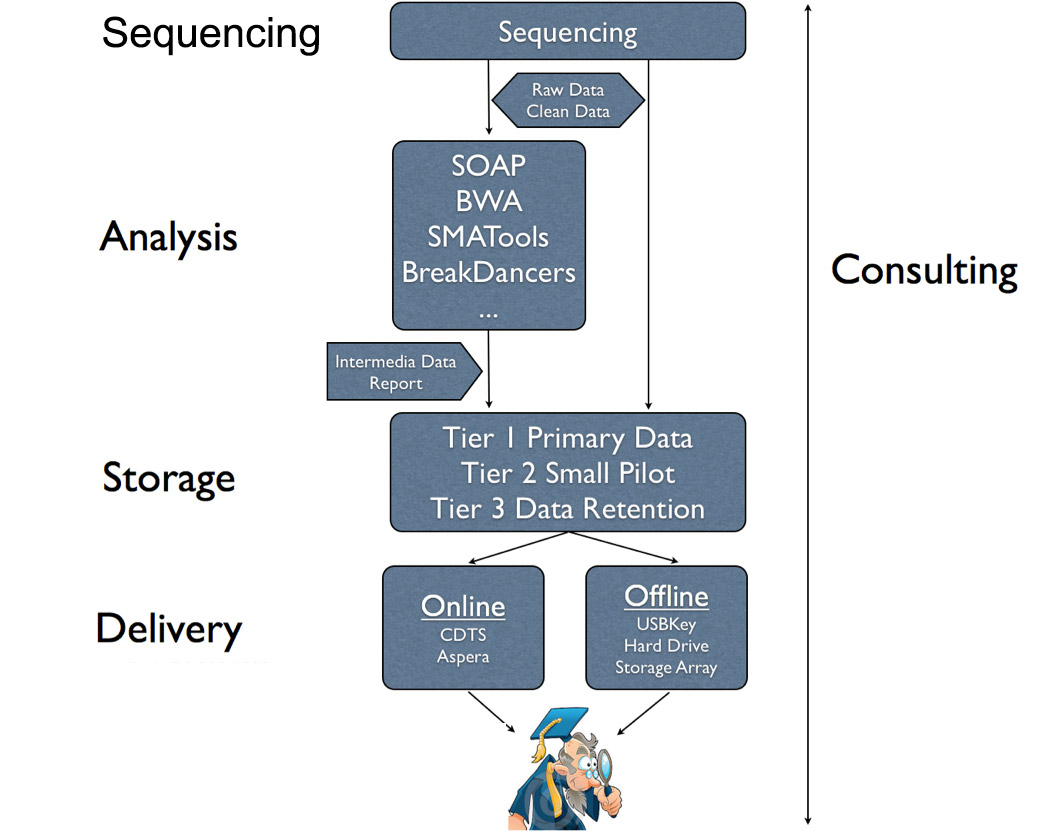 Figure 2. BGI Bio-Cloud Workflow
Figure 2. BGI Bio-Cloud Workflow
Cloud-Based Software in BGI
BGI has developed two new cloud-based software-as-a-service offerings for next-gen data analysis called Hecate and Gaea. They are “flexible computing” solutions for de novo assembly and genome re-sequencing.
Hecate
Hecate is based on a series of distributed algorithms on map/reduce framework to recognize and simplify non-branching repeat-free regions of the genome, correct errors and resolve the ambiguous bubbles and short repeats. Together with the distributed graph shrinkage algorithms to construct a linear DNA sequence, Hecate offers greatly reduced running time and hardware cost for short sequence assembly. For example, using 96 Hecate cores, the genome coverage would increase to 84% in 42 hours at a price of $60,000, a savings of 28 hours and $90,000 compared to SOAPdenovo running on a single server (Table 1).
| Human Genome | SOAP de novo | Hecate (48 cores) | Hecate (96 cores) |
| Genome Coverage | 80% | 84% | 84% |
| Contig N50 | 1,050 | 940 | 940 |
| Contig N90 | 205 | 100 | 100 |
| Scaffold N50 | 440,000 | 300,000 | 300,000 |
| Scaffold N90 | 78,000 | 20,000 | 20,000 |
| Mismatch Rate | 10-6 | 10-6~10-5 | 10-6~10-5 |
| Assembly Time | 70 Hours | 76 Hours | 42 Hours |
| Number of Servers | 1 | 6 | 12 |
| Memory Size | 500 GB x 1 | 24 GB x 6 | 24 GB x 12 |
| Hardware Price | USD 150,000 | USD 30,000 | USD 60,000 |
| Mode | Centralized | Distributed | Distributed |
Table 1. The performance comparisons of Hecate & SOAPdenovo
Gaea
Gaea is designed to distribute re-sequencing computation to a cluster of nodes based on the Hadoop Streaming framework with personalized algorithm interfaces for running the SOAP2, BWA, SAMTools, SOAPsnp, DIndel, and BGI’s realSFS. Gaea is designed to distribute re-sequencing computation to a cluster of nodes to greatly reduce the running time and hardware cost. For the current version of Gaea (v 1.2), speed increases of 75x for SOAP2 and 90x for BWA using 100 cores. At 400 cores those numbers rose to 300x and 346x speed increases compared to running either algorithm on a single core.
| Time (1 core) |
Speedups (100 cores) |
Speedups (200 cores) |
Speedups (400 cores) |
|
| SOAP2 | 82,200s | 75x | 152x | 300x |
| BWA | 62,000s | 90x | 168x | 346x |
| Samtools | 360s | 80x | 155x | 300x |
| DIndel | 46,000s | 60x | 112x | 210x |
| realSFS | 4,000s | 40x | 75x | 140x |
| Hardware Expense | $400 | $40,000 | $80,000 | $160,000 |
| Mode | Centralized | Distributed | Distributed | Distributed |
Table 2. The performance of Gaea software.

