
- Genomics
- Transcriptomics
- Epigenomics
- Meta-omics
- Proteomics
- Single-Cell Sequencing
- Immune Repertoire Sequencing
- FFPE Samples

cDNA Library Construction & EST Sequencing
A cDNA library is a combination of cloned cDNA fragments inserted into a collection of host cells, which together constitute some portion of the transcriptome of the organism. cDNA is produced from fully transcribed mRNA found in the nucleus and therefore contains only the expressed genes of an organism.
A cDNA library can specifically reflect the protein coding gene that is expressed on a particular developmental stage in certain tissues or cell. As such, the cDNA library has a tissue or cell specificity.
Currently, the library constructed includes a full-length cDNA library and full-length normalized cDNA library.
Applications:
- New gene cloning;
- Analysis of gene expression profiles;
- Revealing SSR markers from an EST database;
- Exploring key genes related to disease prevention and development function;
- Performing transgenic function research to change a creature’s characters;
- An EST can be used as probes to construct the molecular marked linkage map.
Workflow:
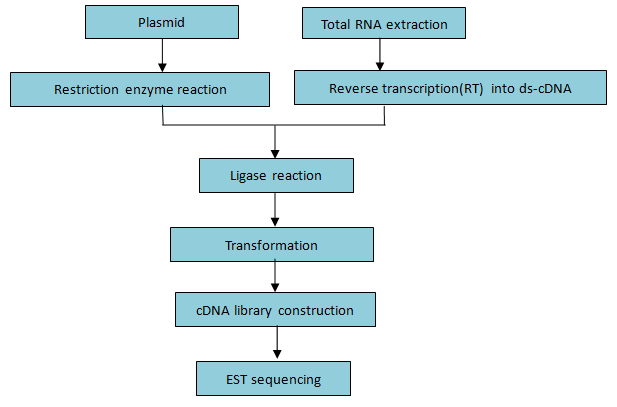 Figure 3. Workflow of cDNA library construction and EST sequencing
Figure 3. Workflow of cDNA library construction and EST sequencingBioinformatics Contents:
cDNA library
- The main results of library qualification include storage capacity, insert segment size, the full length ratio, redundancy rate, etc.
EST sequencing
- Data filtering includes removing adaptors, contamination, and low quality reads from raw reads.
Standard bioinformatics analysis
- Clustering and assembling EST into unigene
- Revealing Unigene’s open reading frame (ORF)
- Revealing the codon, GC content, and read length in each EST database
- Gene expression abundance analysis; unigene splicing, function prediction (Blstx, Blastn ), and classification
Sample Requirements:
- Library Construction Sample Requirements:
Tissue samples:
Please provide a sufficient amount of fresh samples (preserved at 4°C and no older than 1 day or stored in liquid N2 or -80°C and no older than one month). Plant tissue samples should be fresh etiolated seedlings.
RNA samples:
Total RNA should be ≥ 50μg, and RNA content will increase according to the capacity of library; RNA concentration should be > 0.3μg/μL and OD260/280 = 2.0.
- Sequencing Sample Requirements:
1) Use transformed strains
2) Spread the plate overnight, shaking cultivated bacterial strains in a 96-well plate (single bacterial strains). Use streak or stab cultured strains (only aimed at BAC/Fosmid monoclonal sequencing samples).
Turnaround Time:
The standard turnaround time for library construction using the workflow (above) is approximately 30 working days;
The standard turnaround time for EST sequencing using the workflow (above) is approximately 30 working days.
Completion Milestone and Results Submission:
Completion Milestone:
- For cDNA library construction:
The average length of an insert fragment is > 1Kb. (The criterion is the electrophoresis result of 30 colony PCR.) The capacity of the library is > 1×106 clones; the empty clones rate is < 15%.
For a full length cDNA library, the full length ratio of an insert fragment is > 35%;
For a full length normalized cDNA library, the full length ratio of an insert fragment is > 35%, with a redundancy rate of < 10%. - For EST sequencing:
Single reads are ≥ 100 bp, average reads are ≥ 550 bp, and all data include vectors.
Results Submission:
For library construction projects, all ligation products, transformed bacterial liquid, the library check report, and the library appraisal report are included;For sequencing projects, plasmids, clones, raw data, and the data analysis report are included.
