- Genomics
- Transcriptomics
- Epigenomics
- Meta-omics
- Proteomics
- Single-Cell Sequencing
- Immune Repertoire Sequencing
- FFPE Samples

Human
Cases
Technical Information
Contact Us / Wish List
 The ability of NGS to sequence the whole genome has enabled large-scale comparative studies to be performed in an effort to understand how genetic differences affect health and diseases. BGI uses state-of-the art sequencing and bioinformatics technology to achieve a comprehensive analysis of genomic variations. To date, BGI has sequenced more than 16,000 human whole genomes. BGI’s whole genome sequencing can be used to study human genetics, population and evolution, human disease and clinical research, and pharmacogenomics.
The ability of NGS to sequence the whole genome has enabled large-scale comparative studies to be performed in an effort to understand how genetic differences affect health and diseases. BGI uses state-of-the art sequencing and bioinformatics technology to achieve a comprehensive analysis of genomic variations. To date, BGI has sequenced more than 16,000 human whole genomes. BGI’s whole genome sequencing can be used to study human genetics, population and evolution, human disease and clinical research, and pharmacogenomics.
Benefits:
- Deep experience in human whole genome sequencing.
- Industry leading turnaround time
- Affordable, competitive cost
- Reliable, bioinformatics analyses (Mendelian disorder, complex disease, cancer, population analysis)
- Advanced customized analyses with accessibility to deep data sets
Customer Testimonials:
"Knome has been working with BGI for well over three years. Specifically, BGI has been resequencing whole human genomes and exomes for us and it is work they perform exceedingly well. Partnering with BGI has been a positive experience for us and we highly recommend them." - Ari Kiirikki, VP of Knome
Detection of de novo Germline Mutations Hotspots in Autism Spectrum Disorder

De novo mutation plays an important role in autism spectrum disorders (ASDs), which is estimated 1 in 110 children in the U.S. Autism is characterized, in varying degrees, by social and behavioral challenges, as well as repetitive behaviors. The study investigated global patterns of germline mutation by whole genome sequencing of monozygotic twins concordant for ASD and their parents. We found that hypermutability was a characteristic of genes involved in ASD and other diseases. The findings suggest that regional hypermutation is a significant factor in shaping patterns of genetic variation and disease risk in humans.
The Netherlands Genome Project

The Project will establish a map of genetic variants in 750 samples from Dutch biobanks and build the reference database for future disease oriented genomics research. BGI is responsible for sequencing 750 whole genomes representing 250 trios (the genomes of two parents and one offspring) which will help to determine how a certain variant fits within the paternal and maternal patterns. This project is expected to reveal most of the genetic variation within the Dutch population and among different regions. It is expected that this study will reveal many low-frequency variants that were unobserved in the European 1000 Genomes data.
Ancient Human Genome Sequence of an Extinct Palaeo-Eskimo.

In 2010, BGI and Copenhagen University obtained DNA from a 4,000-year-old permafrost-preserved human hair, and isolated and sequenced its genomic DNA. The genome is from a male individual who was a member of the first known population to settle in Greenland. SNP analysis of the sequence data enabled the researchers to assign possible phenotypic characteristics of this individual and identify the population to which he is most closely related. This analysis provided evidence that there was human migration from Siberia into the New World about 5,500 years ago, and this migration was independent of the one that gave rise to the modern Native Americans and Inuits.
The Diploid Genome Sequence of an Asian Individual.
Nature 2008; 456 (7218): 60-65.
 The first Asian individual genome was sequenced for 36X by massively parallel sequencing technology. The NCBI human reference genome was covered at 99.97%. And the resulting high-quality consensus sequence represents 92% of the individual genome. The data quality and analyses demonstrated the potential usefulness of next-generation sequencing technologies for personal genomics.
The first Asian individual genome was sequenced for 36X by massively parallel sequencing technology. The NCBI human reference genome was covered at 99.97%. And the resulting high-quality consensus sequence represents 92% of the individual genome. The data quality and analyses demonstrated the potential usefulness of next-generation sequencing technologies for personal genomics.
Other Publications:
A Map of Human Genome Variation from Population-Scale Sequencing. Nature. 27 Oct 2010; 467: 1061-73.
Bioinformatics:
Comparison of different genome sequences allows for the detection and annotation of genomic variations, including single nucleotide polymorphisms (SNPs), insertions and deletions (indels), and various structural variations (SVs). In addition, re-sequencing a variety of genomes allows evolutionary analyses to be performed at the population level.
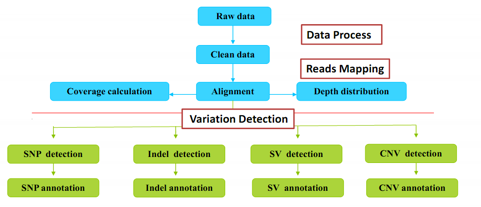
BGI provides the following bioinformatics analysis.
Standard Analysis
- Data filtering (removing adaptors, contamination, and low-quality reads from raw reads)
- Alignment and result report
- SNP calling, annotation and statistics
- Indel calling, annotation and statistics
- Copy number variation (CNV) calling, annotation and statistics
- Structural variation (SV) calling, annotation and statistics
Advanced Analysis
- Mendelian disorders advanced bioinformatics analysis
- Complex diseases advanced bioinformatics analysis
- Cancer advanced bioinformatics analysis
- Population analysis
Custom Analysis
- We also offer customized bioinformatics analysis based on your specific research needs.
Sample Requirements:
- Purity: OD260/280=1.8-2.0 without degradation and RNA contamination
- Concentration: ≥25ng/μl
- DNA amount: ≥1 μg (although more than 2.5 μg is recommended)
Recommended Sequencing Depth:
- For individual research, the recommended sequencing depth is about 30X.
- For population research, the recommended sequencing depth is 5X~15X.
Turnaround Time:
The standard turnaround time for the workflow (above) is about 40 business days.
Technologies:
BGI's current sequencing capacity is 10 Tb a day (equivalent to 3333X coverage of a human genome per day).

