- Genomics
- Transcriptomics
- Epigenomics
- Meta-omics
- Proteomics
- Single-Cell Sequencing
- Immune Repertoire Sequencing
- FFPE Samples

Whole Genome Mapping
Cases
Technical Information
Contact Us / Wish List
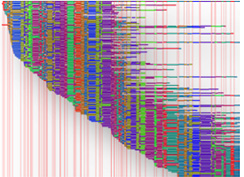 A Whole Genome Map is a high-resolution, ordered, whole genome restriction map generated from single DNA molecules extracted from bacteria, yeast, or other fungi. Whole Genome Mapping is a novel technology with unique capabilities in the field of microbiology, with specific applications in the areas of Comparative Genomics, Strain Typing, and Whole Genome Sequence Assembly. Whole Genome Maps are generated de novo, independent of sequence information, require no amplification or PCR steps, and provide a comprehensive view of whole genome architecture. A Whole Genome Map is displayed in the MapCode pattern where the vertical lines indicate the locations of restriction sites, and the distance between the lines represent the restriction fragment size.
A Whole Genome Map is a high-resolution, ordered, whole genome restriction map generated from single DNA molecules extracted from bacteria, yeast, or other fungi. Whole Genome Mapping is a novel technology with unique capabilities in the field of microbiology, with specific applications in the areas of Comparative Genomics, Strain Typing, and Whole Genome Sequence Assembly. Whole Genome Maps are generated de novo, independent of sequence information, require no amplification or PCR steps, and provide a comprehensive view of whole genome architecture. A Whole Genome Map is displayed in the MapCode pattern where the vertical lines indicate the locations of restriction sites, and the distance between the lines represent the restriction fragment size.
Benefits:
With Whole Genome Mapping, you will be able to investigate microbial structure, function, diversity and genetics— without the need for amplification, PCR, cloning, paired-end libraries, pure isolates, or genomic specific reagents. Using OpGen’s unique de novo Whole Genome Mapping Technology, the Argus Whole Genome Mapping System, BGI delivers high resolution, ordered whole genome restriction maps from single microbial DNA molecules.
Applications:
- Comparative Genomics
- Whole Genome Sequencing Assembly
- Strain Typing
Case 1: Assisting Bacteria Genome Assembly - The Aacinetobacter baumannii Case
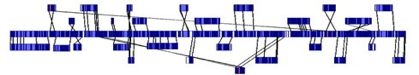
In this case, unordered contigs were aligned to the matching regions on the Whole Genome Map. Alignment lines are drawn between compared Whole Genome Maps to show placement. Crossing alignment lines indicate reverse orientation. The Gap sizes and locations (white colored) are visualized and enable further targeted analysis for whole genome closure.
Comparison of Assembly Performance Among Three Methods:
Method 2 combines Whole Genome Mapping and Solexa reads assembly, bringing the number of scaffolds to 1. It works better than method 1 and 3 which use large and small fragments to assemble without Whole Genome Mapping.
| Method | Number of scaffolds | Scaffold total length | Total length of outer gaps and inner gaps | Contig length/ genome length (%) |
| Assembly (short library reads only) | 88 | 4,060,072 | 309,996 | 85.88% |
| Assembly (short reads) + Whole Genome Mapping | 1 | 4,292,545 | 426,960 | 90.05% |
| Assembly (short reads + longer reads) | 74 | 4,366,732 | 307,400 | 92.96% |
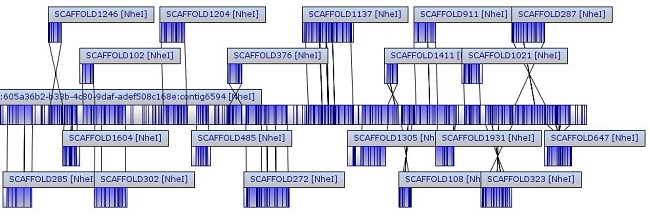
Case 2: Optical Mapping for a Fungus
The genome was assembled into seven chromosomes, with total size of 37.2 Mb after finishing and rescaling (similar to the 38.0 Mb sequence genome). All seven chromosomes passed assembly QC, ensuring high quality results.

245 of 292 sequence scaffolds (> 40 kb) were successfully placed on optical maps, with a placement rate of 84%. The total size of the placed scaffolds is 27.4 Mb (out of 29.87 Mb). The unplaced scaffolds tend to be small and have small numbers of cut sites. The placed scaffolds show a very high level of agreement with optical maps.
Bioinformatics:
For animal or plant samples
- Enzyme digestion result
- Assembly and analysis
- Scaffold placement
For bacterial samples
- Enzyme digestion result
- Assembly and analysis
- Optical map
- Scaffold placement
- Comparative genomics
- Strain typing
Sample Requirements:
For animal or plants
- Sample size: the genomic DNA Band should be ≥250 kb
Note: We recommend that you send your tissues to BGI Shenzhen and we extract DNA for you
For bacteria
- Sample condition: Bacteria DNA with size≥150Kb
We suggest sending bacterial cultural plate. For bacterial cultural plate, two more control plates are needed for re-culture. Label the culture condition and pathogenicity.
For pathogenic bacteria, we suggest sending Bacteria Plug.
Turnaround Time:
For animal or plant samples, the standard turnaround time is approximately 50 business days.
For bacterial samples, the standard turnaround time is approximately 5 business days.

