
- Genomics
- Transcriptomics
- Epigenomics
- Meta-omics
- Proteomics
- Single-Cell Sequencing
- Immune Repertoire Sequencing
- FFPE Samples

Yeast Two-hybrid Library Construction
The establishment of the yeast, two-hybrid library is based on the understanding of the regulation of the transcription initiation process of eukaryotic cells. Many eukaryotic transcription activation factors come from two separate, functional, independent domains. For example, the yeast transcriptional activation factor GAL4 in the N terminal has a DNA binding domain and is composed of 147 amino acids (DNA binding domain (BD)). However that factor in the C terminal has a transcriptional activation domain and is composed of 113 amino acids (transcription activation domain (AD)). The DNA binding domain of GAL4 can bind with upstream activation sequence (upstream activation sequence (UAS)), whereas the transcriptional activation domain can activate UAS downstream genes to transcription. However, a separate DNA binding domain cannot activate gene transcription, and a separate transcription activation domain cannot activate the UAS downstream genes. The DNA binding domain and transcription activation domain have to be combined to derive the complete function of the transcription activation factor.
The yeast two-hybrid library can be used to study the interaction between known proteins, to find to the domain that plays a key role in protein-protein interaction and to discover a new protein interaction with target proteins.
The yeast two-hybrid library has the following applications:
1) Interactions between new proteins and protein identification;
2) Protein cascade substrate identification;
3) Effects of mutations on protein and protein binding identification;
4) Identification of interfered protein among known interactions (reverse two-hybrid system).
Workflow:
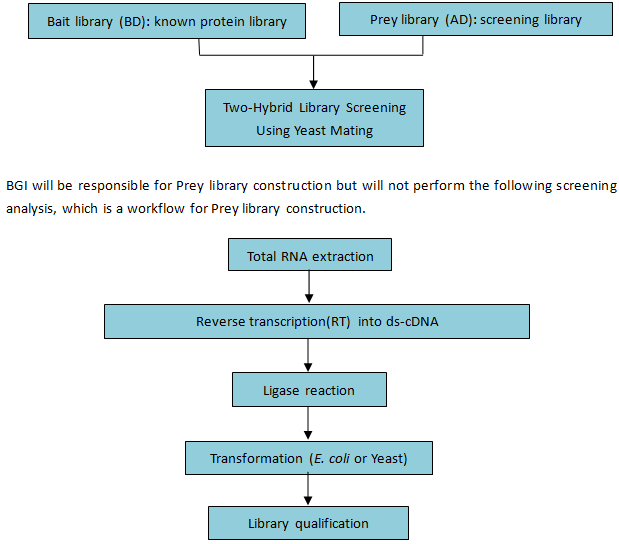 Figure 4. Workflow of yeast two-hybrid library construction
Figure 4. Workflow of yeast two-hybrid library constructionBioinformatics:
Transformed E. coli: The main results of library qualification include storage capacity, insert segment size, the full length ratio, redundancy rate, etc.
Transformed yeast: The main results of library qualification include storage capacity, insert segment size, etc.
Sample Requirements:
Tissue samples:
Please provide a sufficient amount of fresh samples (preserved at 4°C and no older than 1 day or samples stored in liquid N2 or -80°C and no older than one month). Plant tissue samples should be fresh etiolated seedlings.
RNA samples:
Total RNA should be ≥ 50μg, and RNA content will increase according to the capacity of the library. RNA concentration should be > 0.3μg/μL; and OD260/280 = 2.0.
Turnaround Time:
For a full-length library, the standard turnaround time using the workflow (above) is approximately 30 working days;
For normalized library, standard turnaround time for the workflow (above) is approximately 40 working days.
Completion Milestone and Result Submission:
Transformed E. coli
Completion milestone:
The average insert fragment size should be 1 kb. The capacity of the library should be > 1×106 clones and the empty clones rate should be < 15%;
For full length library, the full length ratio of the insert fragment should be > 35%;
For a normalized library, the redundancy rate should be < 10%.
Results submission:
Results submitted include transformed bacterial liquid (initial capacity of library > 1×106), the sequencing library check report, and the final report.
Transformed yeast
Completion milestone:
The average insert fragment size should be 1kb and the capacity of library > 1×106clones. The empty clones rate should be <15%.
Results submission:
Results include the transformed bacterial liquid (initial capacity of library >1×106 ) and the final report.
