 Human cancers typically carry several genomic rearrangements, such as copy number alteration
Human cancers typically carry several genomic rearrangements, such as copy number alteration s and point mutations. These genetic alterations are important diagnostic, prognostic, and predictive biomarkers to help design personalized treatments thatare more effective and less toxic.
s and point mutations. These genetic alterations are important diagnostic, prognostic, and predictive biomarkers to help design personalized treatments thatare more effective and less toxic.
BGI TumorCareTM is a newly introduced NGS-based panel solution designed to detect all types of tumor genomic alterations, including base substitutions, insertions and deletions, copy number alterations, and selected fusions. BGI, leveraging our years of cancer research experience combined with extensive literature and database searches, has designed a comprehensive cancer gene panel. BGI TumorCareTM includes 1053 genes that (1) are strongly supported as clinically-actionable cancer genes, (2) harbor high frequency alterations in the COSMIC database, or (3) play key roles in core cancer pathways.
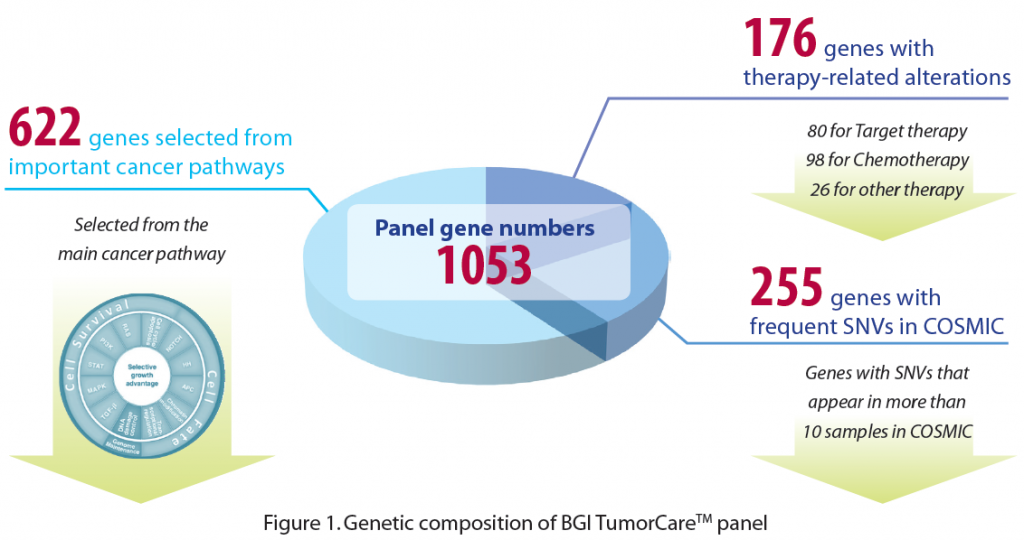
Followed by target region sequencing and our bioinformatics analysis pipeline, all types of alterations are detected with high sensitivity and specificity (Table 1). Moreover, using our extensive in-house cancer knowledgebase, including Drug-Bank database, PharmaGKB database, and cancer research literature, this panel allows researchers to identify and investigate the associations between detected genomic alterations and 117 known cancer therapeutics, including those approved by FDA and in clinical trials. This knowledge may help us understand more about the effects and cytotoxicity of specific treatments, such as target therapy and chemotherapy.
Benefits:
• Comprehensive detection: SNVs, INDELS, CNVs, and gene fusions of 1053 genes in a single assay for a wide variety of cancer types
• High sensitivity and specificity: >99% in ≥1% minor allele frequencies (Table 1)
• Low input DNA amounts as low as 200 ng
• Enables in-depth investigation of genetic variations and their associations to target therapies and traditional chemotherapies through our extensive knowledgebase.
• Experienced team: Our team has analyzed tens of thousands of cancer samples and has many publications in top-tier journals
• Affordable cost per sample: Contact us to learn more.
• FFPE, normal tissue, biopsy, and single-cell samples supported
Table 1. Accurate and sensitive detection of mutations
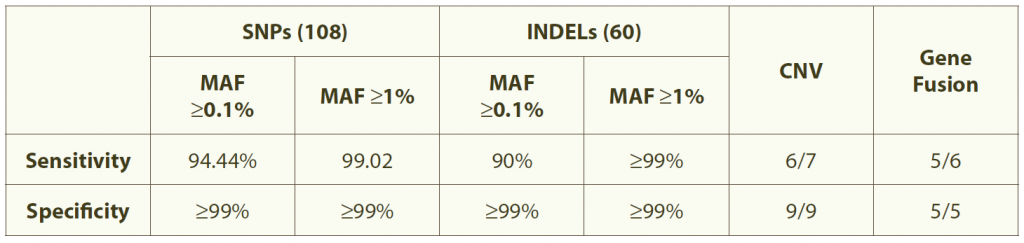
Note: Data are from standard tumor samples with known alterations (108 SNPs and 60 INDELS) without matched normal tissues. Sequencing was performed using HiSeq200 pair-end 101 with an initial DNA amount of 200 ng.
For research use only. Not for therapeutic or diagnostics use.

