
- MHC-Seq: Capturing Sequencing and Typing of MHC Regions New!

- Human RRBS Promotion 50% Off
The promotion is valid for Americas and Europe customers only NOW through MAY 31!
Human MHC-Seq: Capturing Sequencing and Typing of MHC Regions New!
Major Histocompatibility Complex (MHC) plays a critical role in hundreds of diseases and is a key target in drug R & D. BGI and Roche NimbleGen co-developed a MHC region capture technology to allow easy capture and enrichment, and deep sequencing of these highly repetitive regions. The technology targets the MHC region (3.37MB) and approximately 1.6Mb of the regions surrounding MHC, providing a total of 4.97Mb. Not only does this technology enable accurate genotyping of known SNPs (with a 99.42% concordance with traditional genotyping technologies), it provides the opportunity to discover novel SNPs.
Promotion Details (Valid from NOW through May 31)
The 499 USD/sample package includes:
- Capturing of MHC and surrounding regions: The technology not only targets the traditional MHC region (3.37MB), but also targets approximately 1.6Mb of the regions surrounding MHC, providing a total of 4.97Mb.
- 90 bp pair-end sequencing on HiSeq 2000
- 80X high quality mapping data guaranteed
- SNP and Indel calling and annotation
- Typing of 26 HLA genes at 4 digit resolution with 95% accuracy
- ~97% of MHC and extended regions can be covered by sequencing reads.
Minimum order: 30 samples. The first batch of samples must be sent to BGI before May 31.
Turnaround time: 6-8 weeks for up to 400 samples
Contact us to get started!
Promotion of RRBS 50% off (only for human and mouse)
RRBS is a high-throughput, cost-effective method for genome-wide DNA methylation studies. RRBS enriches CpG-rich regions, such as CpG islands and promoter regions, thereby focusing the sequencing efforts over regions of interest. The approach provides single-nucleotide resolution, is highly sensitive and is suitable for clinical samples.
A. 3G high-quality data
- 999 USD per sample, including library construction, sequencing and standard bioinformatics analysis.
- 50 bp paired-end sequencing on HiSeq 2000.
- Cover above 50% of the CpG islands and promoter regions.
- ~82% and ~61% CpG sites covered with sequencing depth >=4X and 10X, respectively.
B. 5G high-quality data
- 1299 USD per sample, including library construction, sequencing and standard bioinformatics analysis.
- 50 bp paired-end sequencing on HiSeq 2000.
- Improve the coverage by ~2% of the CpG islands and promoter regions compared to that of 3G high-quality data.
- ~85% and ~70% CpG sites covered with sequencing depth >= 4X and 10X, respectively.
- High reproducibility at different coverage depths
*High-quality data are generated from raw data with adaptor and low quality sequences removed.
Standard bioinformatics analysis includes:
- Adapter pollution removal, low quality reads filtering, and production statistics
- Alignment of RRBS sequence to the reference
- Coverage analysis of promoter and CGI
- Methylation analysis of promoter and CGI
- DMR analysis
We can also perform custom analysis to meet your specific requirements.
Why is the 5G high-quality data more suitable for performing biological replicates for multiple samples from the same species?
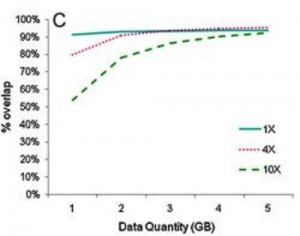 Fig. 1 The proportion of the same CpG sites covered by two biological replicates at different coverage depths with increasing size of data set. For 3G high-quality data, the proportion of the same CpG sites covered by two biological replicates may reach ~90% at 1X and 4X sequencing depths and reach ~83% at 10X sequencing depth. For 5G high-quality data, the proportion of the same CpG sites covered by two biological replicates may reach ~90% at 1X, 4X and 10X sequencing depths, which indicates good reproducibility at different coverage depths and is more suitable for performing biological replicates for multiple samples from the same species. Fig. 1 The proportion of the same CpG sites covered by two biological replicates at different coverage depths with increasing size of data set. For 3G high-quality data, the proportion of the same CpG sites covered by two biological replicates may reach ~90% at 1X and 4X sequencing depths and reach ~83% at 10X sequencing depth. For 5G high-quality data, the proportion of the same CpG sites covered by two biological replicates may reach ~90% at 1X, 4X and 10X sequencing depths, which indicates good reproducibility at different coverage depths and is more suitable for performing biological replicates for multiple samples from the same species. |
Minimum order: 10 samples. The first batch of samples must be sent to BGI before May 31.
Turnaround time: 7-9 weeks for up to 20 samples
Minimum DNA quantity: 1 μg (5 μg for two library constructions are recommended)
BGI Publication: Wang, Li, et al. Systematic assessment of reduced representation bisulfite sequencing to human blood samples: A promising method for large-sample-scale epigenomic studies. Journal of Biotechnology 151(1), 1-6 (2012).
Contact us to get started!


