
de novo Sequencing
Cases
Technical Information
Contact Us / Wish List
 de novo sequencing provides the first genome sequence of an organism. With the advent of rapid, low-cost next-generation sequencing technology researchers can now obtain whole genome data for organisms previously considered too low a priority to sequence. The availability of this whole genome data has allowed large-scale genomic studies to be performed that were unimaginable just a few years ago. BGI and its global collaborators have now initiated 580 plant and animal genome projects, and among them, completed fine or draft genome maps for nearly 150 species. The completed projects include rice, silkworm, cucumber, panda, camel, oyster, ant genomes, and more.
de novo sequencing provides the first genome sequence of an organism. With the advent of rapid, low-cost next-generation sequencing technology researchers can now obtain whole genome data for organisms previously considered too low a priority to sequence. The availability of this whole genome data has allowed large-scale genomic studies to be performed that were unimaginable just a few years ago. BGI and its global collaborators have now initiated 580 plant and animal genome projects, and among them, completed fine or draft genome maps for nearly 150 species. The completed projects include rice, silkworm, cucumber, panda, camel, oyster, ant genomes, and more.
Benefits:
- More comprehensive maps of genetic variation
- Variable gradient insert libraries enable fine mapping of the genome
- Reliable genome assembly by BGI’s independently developed software- SOAPdenovo
- NGS high-throughput sequencing reduces cost
- Experienced bioinformatics team
The Sequence and de novo Assembly of the Giant Panda Genome. Nature 2010.463:311-317.

The panda genome was the first genome completely sequenced by using next generation sequencing platform alone. It provides clues to the understanding of everything from the panda’s strict bamboo diet to it’s genetic diversity. It may also aid in the panda conservation in the future.
Genome Sequence and Analysis of the Tuber Crop Potato. Nature. 14 Jul 2011; 475:189-195.

Potato (Solanum tuberosum L.) is the world’s most important non-grain food crop and is central to global food security. It is clonally propagated, highly heterozygous, autotetraploid, and suffers acute inbreeding depression. Here we use a homozygous doubled-monoploid potato clone to sequence and assemble 86% of the 844-megabase genome. We predict 39,031 protein-coding genes and present evidence for at least two genome duplication events indicative of a palaeopolyploid origin.
The Genomic Sequence of the Chinese Hamster Ovary (CHO)-K1 Cell Line. Nature Biotechnology. 31 Jul 2011; 29: 735–741.

Chinese hamster ovary (CHO) cell lines are the preferred and most commonly used mammalian cell line today in biological and medical research. With the advanced genome sequencing capability of BGI, the CHO-K1 ancestral cell line was sequenced by de novo sequencing and then assembled by BGI's Short Oligonucleotide Analysis Package, SOAPdenovo, resulting in the assembly of 2.45 Gb of the genomic sequence. This information was combined with transcriptome sequence data and led to the generation of 24,383 predicted genes. The study yields a better understanding of the genetics of CHO cells and will accelerate the discovery and development of new recombinant protein therapies.
Structural Variation in Two Human Genomes Mapped at Single-Nucleotide Resolution by Whole Genome de novo Assembly. Nature Biotechnology. 2011.29:723-730.
Whole genome de novo assembly was used to map structural variations in an Asian and an African genome. Small-and-intermediate-size homozygous variants including insertions, deletions, and inversions were identified. These findings also demonstrate that whole genome de novo sequencing is a practical approach to deriving more comprehensive maps of genetic variation.
Workflow: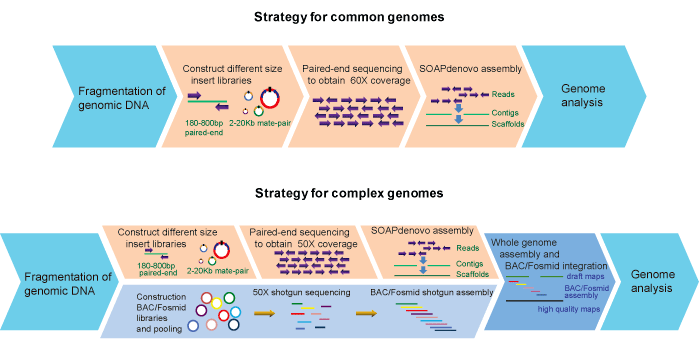
Bioinformatics:
|
Assembly |
|
|
Annotation |
|
|
Evolution analysis for animal and plant species |
|
|
Advanced bioinformatics for microbial species |
|
Sample Requirements
- Sample quantity required (single pair):
- Short-insert libraries: ≥3 µg
- 2 kb large-insert libraries: ≥20 µg
- 5 kb-6 kb large-insert libraries: ≥20 µg
- 10 kb large-insert libraries: ≥30 µg
- 20 kb and 40 kb large-insert libraries: ≥60 µg
- PCR-free libraries with high or low GC content: ≥30 µg
Note: the total sample quantity required is also determined by the experimental strategy, as well as the type and number of libraries to be constructed.
- Sample concentration:
- Short-insert libraries: ≥30 ng/ µL
- Large-insert libraries: ≥133 ng/ µL
- Sample quality: genomic DNA should be intact.
- Sample purity: OD260/280= 1.8-2.0
Turnaround Time:
| Animals/Plants | Survey: 2 months from sample qualification | Common genome: 6 months from sample qualification | Complex genome: 12 months from sample qualification |
| Fungi | Survey: 40 business days | Draft map: 50 business days | Fine map: 50 business days (from completion of survey) |
| Bacteria | Survey: 40 business days | Fine map: 60 business days | Complete map: 75 business days |
Completion Criteria
Genomic map of plant or animal species
| Genome Size (GS) | Assembly Indicator |
| ≥ 300 Mb | Contig N50 > 20 kb; Scaffold N50 > 300 kb |
| Contig N50 > 10 kb; Scaffold N50 > 150 kb | |
| 300 Mb < GS ≤ 1500 Mb (except birds) | Contig N50 > 20 kb; Scaffold N50 > 300 kb |
| Contig N50 > 10 kb; Scaffold N50 > 150 kb | |
| 1500 Mb < GS ≤ 3000 Mb (except mammals) | Contig N50 > 10 kb; Scaffold N50 > 150 kb |
| Contig N50 > 5 kb; Scaffold N50 > 20 kb | |
| GS < 1600 Mb (birds) | Contig N50 > 20 kb; Scaffold N50 > 300 kb |
| GS < 3200 Mb (mammals, except Chiroptera) | Contig N50 > 20 kb; Scaffold N50 > 300 kb |
| Complex genomes | Contig N50 > 20 kb; Scaffold N50 > 300 kb |
Genomic map of microbial species
| Fungi | Survey | Sequencing depth ≥ 30X |
| Draft map | Sequencing depth ≥ 50X | |
| Fine map | The coverage of chromosome or chromatin genome is > 95%.The coverage of a gene region is > 98%Scaffold N50 > 300 kb, with an overall sequencing depth ≥ 50X. | |
| Bacteria | Survey | Sequencing depth ≥ 100X |
| Fine map | The coverage of chromosome or chromatin genome is > 95%.The coverage of a gene region is above 98%The overall sequencing depth is ≥ 100X. | |
| Complete map | Provide 1 contig sequence and PCR validation. | |
| Small genome | Survey | Sequencing depth ≥ 100X |

