Monkey Exome Sequencing
Cases
Technical Information
Contact Us / Wish List

Due to its close relationship genetically and physiologically to humans, the monkey is the most extensively used non-human primate in biomedical research and animal models for human disease research. Macaque monkeys such as rhesus macaques, in particular, are commonly used in research. Each year, there are thousands of Macaques used in pre-clinical studies throughout the world, including studies of pathogenic mechanisms, drug selection, drug dosage, treatment duration and adverse drug reactions, among others.
BGI has developed the first exome sequencing platform for the monkey, based on next-generation sequencing technology and monkey exome capturing array (MECA). MECA is a proprietary exome capture array designed by BGI for capturing the entire monkey exome. The combination of this revolutionary array and BGI’s high-throughput sequencing technology not only can simplify the workflow of exome sequencing experiments, but also improve cost-effectiveness and turnaround time.
Benefits
- Highly effective: Designed towards specific species; more effective for capturing the monkey exons
- Wide detection range: Capture both common and rare variants
- Highly cost-effective: Focus on ~1% of the genome where most disease- or trait-related variations are located.
Here we report the results of monkey exome sequencing using MECA, a proprietary exome capture array designed by BGI for capturing the entire monkey exome. Download the following white paper to learn about the results of monkey whole exome sequencing and its application in disease research and drug development.
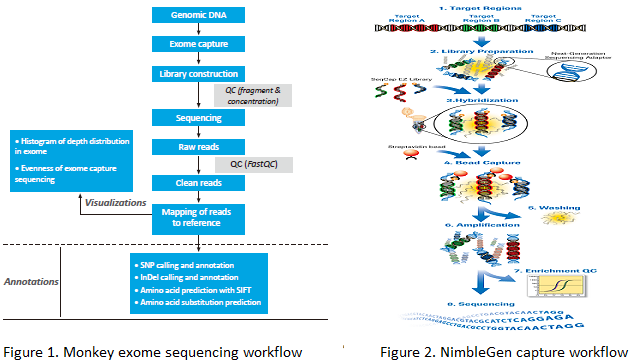
The exome capture array is designed by BGI and produced by Roche NimbleGen. Its capacity is about 50 Mb, covering all exons of rhesus macaque and cynomolgus macaque. The library is prepared using Illumina Paired-End Genomic DNA Sample Prep Kit. Sample and Exome Libraries are hybridized after amplification using LM-PCR. The DNA not complementary to probes is then washed off, and the captured DNA is amplified by LM-PCR again. There is a QC step to measure the enrichment using qPCR and measure the fragments using Agilent 2100 before library construction. Finally, the sequencing data is analyzed by BGI's standard bioinformatics pipeline.
Sample Requirements:
- Purity: OD260/280 =1.8~2.0, without degradation and RNA contamination
- Concentration: ≥ 50-200 ng/μl
- Quantity demanded: ≥ 6 μg (for two times library construction in case there is a failure)


