- Genomics
- Transcriptomics
- Epigenomics
- Meta-omics
- Proteomics
- Single-Cell Sequencing
- Immune Repertoire Sequencing
- FFPE Samples

FFPE Samples
Cases
Technical Information
Contact Us / Wish List
 Formalin-fixed, paraffin-embedded (FFPE) samples are common biological materials for disease diagnosis and scientific research. Because FFPE sample tissues can be stored indefinitely at room temperature, and nucleic acids (both DNA and RNA) can be recovered after decades from the original fixation, they have become an important resource for historical studies in medicine. It is, however, challenging to get intact information from such samples, as severe degradation, damage, and molecular or biological modification could occur during sample preparation and store process.
Formalin-fixed, paraffin-embedded (FFPE) samples are common biological materials for disease diagnosis and scientific research. Because FFPE sample tissues can be stored indefinitely at room temperature, and nucleic acids (both DNA and RNA) can be recovered after decades from the original fixation, they have become an important resource for historical studies in medicine. It is, however, challenging to get intact information from such samples, as severe degradation, damage, and molecular or biological modification could occur during sample preparation and store process.
In recent years, there were many publications about the discovery of gene variations, gene expression profiling changes, and protein biomarkers with FFPE tissues, which generally adopt immunohistochemical method, Real-time PCR, and array-based detection methods. However, there are quite few usages of next-gen sequencing (NGS) method in deciphering the FFPE nucleotide information. In order to unveil the information concealed in FFPE samples, BGI has worked relentlessly in applying NGS to FFPE samples. We expect that NGS technologies with FFPE samples will substantially facilitate our understanding of undefined pathological mechanisms and broaden our insights in biomedical research.
Benefits:
- Low input: As low as 200ng as for WGS, WES and RNA-Seq.
- High throughput: Million of reads available for downstream analysis.
- FFPE-Omics:Comprehensive information with multiple sequencing technologies.
- High rRNA removal efficiency: Lower than 10% of the reads mapping on rRNA gene sequences, leading to increased sample throughput
At BGI, we have successfully conducted many genomics and trancriptomics studies on FFPE samples, including whole genome resequencing, whole exome & target region sequencing, RNA-Seq (Transcriptome), and small RNA sequencing. Moreover, We have generated results with high concordance between FFPE and the corresponding Fresh Frozen (FF) samples.
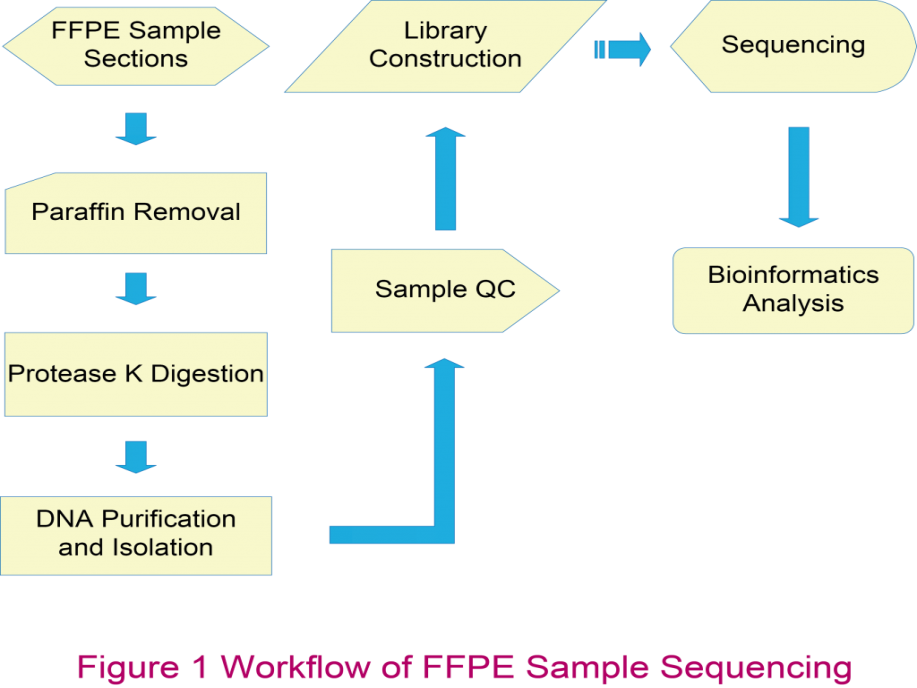
Whole Genome Resequencing
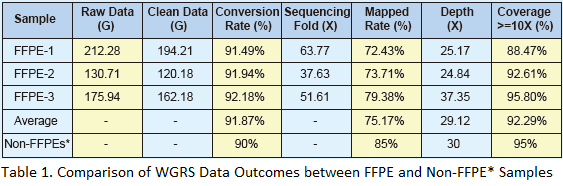
Our whole genome resequencing results suggest that the FFPE derived sequencing data has more than 90% conversion rate (clean data/raw data), indicating that the total DNA isolated with the extraction kits is of sufficient quantity and quality for sequencing using Illumina HiSeq 2000.
Whole Exome Sequencing
Table 2. Comparisons of WES data between FFPE and FF Samples. The data are from exome sequencing with Agilent 50M kit on 50 FF samples and 50 FFPE samples.
To investigate whether information obtained from FFPE samples was reliable, we compared the concordance of single nucleotide polymorphisms (SNPs) between two pairs of tumor tissues and normal controls with two different storage conditions, FFPE and FF. The genome variation pattern obtained from FFPE samples is comparable to that from FF samples (Figure 1).
Figure 1. The concordance of SNPs between FFPE and FF samples (T, Tumor; N, Normal control)
RNA-Seq (Transcriptome)
RNA extracted from FFPE samples is often of poor quality and low yield, which creates difficulties in transcriptome research. BGI has developed highly efficient rRNA depletion methods with Ribonuclease H (RNase H) for library construction on only 100 ng total RNA input of FFPE samples with different storage ages (Figure 2).
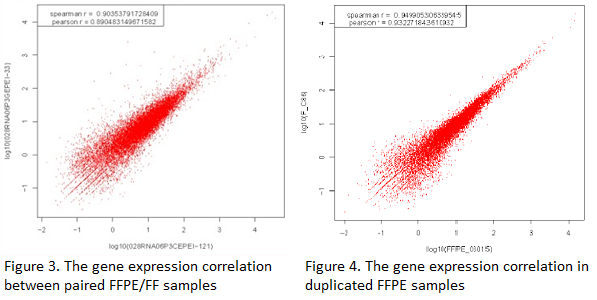
The gene expression correlation between paired FFPE/FF samples and duplicated FFPE samples shows high Pearson R (>0.89), indicating RNA-Seq (Quantification) has a good performance on FFPE samples (Figure 3 and 4).
Small RNA Sequencing
1μg of total RNA from each sample was used for library construction, and clean reads were annotated in different small RNA or mRNA degraded fragments by mapping to different non-coding RNA databases.
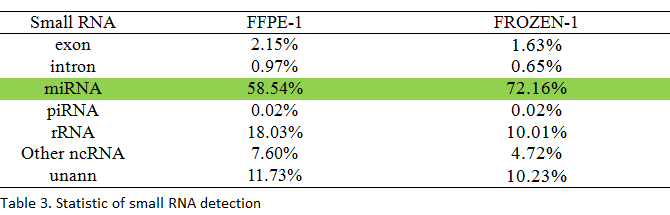
The data suggested that small RNA sequencing is a powerful method for miRNA and piRNA detection, and the results are similar to the ones obtained from the FF samples.
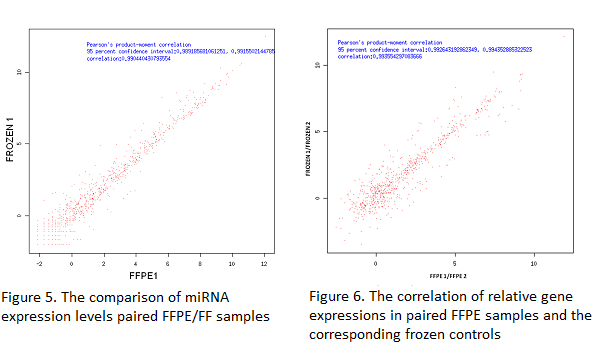
The Pearson ratio is higher than 0.99, which suggests a high technical reproducibility of miRNA detection by small RNA sequencing in FFPE samples and its high correlation to the FF controls.
Other Applications
We are also testing other NGS applications on FFPE samples, including Reduced Representation Bisulfite Sequencing (RRBS) and proteome profiling, which would give researchers more flexibility in the near future. The results would enable full use of the vast number of FFPE samples available for biomedical researches and applications.

Most tumor tissue samples are preserved in the form of FFPE blocks, which in general present several challenges, including variability in fixation methods, diverse ages and sample storage environment, as well as damages that may occur to the nucleic acids during the FFPE process. To overcome the challenges of nucleic acid extraction from FFPE samples and library construction, we have developed a complete and efficient protocol and optimized the conditions at every step in our workflow.
Sample Requirements:
|
DNA-Seq |
FFPE tissue sample |
No less than 8 sections of 10-μm-thick 100 mm2 (tissue area) FFPE tissues are recommended. |
|
gDNA sample (WGRS) |
Input amount: ≥500N ng, N represents the number of constructed library; Concentration: ≥10 ng/ul; Main band size ≥250bp. |
|
|
gDNA sample (WES & Target Region Sequencing) |
Input amount: ≥200 ng; concentration ≥2.5 ng/μl; Main band size ≥500bp OR Input amount: ≥1ug; concentration ≥10 ng/μl; Main band size ≥250bp. |
|
|
RNA-Seq |
FFPE tissue sample |
No less than 5 sections of 10-μm-thick 100 mm2 (tissue area) FFPE tissues are recommended. |
|
Total RNA sample |
Total RNA: ≥200 ng; RNA concentration: ≥20 ng/μl; RIN ≥2.3. |
# The FFPE sample can be stored and delivered under room temperature.