- Genomics
- Transcriptomics
- Epigenomics
- Meta-omics
- Proteomics
- Single-Cell Sequencing
- Immune Repertoire Sequencing
- FFPE Samples

Small RNA Sequencing
Cases
Technical Information
Contact Us / Wish List
 Small RNAs have emerged as important regulators in recent years. Small RNAs have significant functions in many cellular processes including development, cell differentiation and apoptosis, and have been implicated in many diseases. BGI’s small RNA sequencing and bioinformatics analysis services provide reliable detection and profiling of small RNAs rapidly and cost effectively.
Small RNAs have emerged as important regulators in recent years. Small RNAs have significant functions in many cellular processes including development, cell differentiation and apoptosis, and have been implicated in many diseases. BGI’s small RNA sequencing and bioinformatics analysis services provide reliable detection and profiling of small RNAs rapidly and cost effectively.
Benefits:
- Detection of virtually all small RNAs, repeat associated small RNAs, and degraded tags of exons and introns
- A wide range of transcript detection from two to thousands of copies
- Independent of known information, capable of detecting known miRNAs and predicting and detecting novel miRNAs at the same time
- Target gene prediction and function annotation
- High resolution: discriminating of single-base differences
Customer Testimonials:
"BGI offered a good price and the shortest turnaround time… The advanced bioinformatics service helped a lot in the data evaluation… During the process and even after finishing the project, the staff always responded quickly to our questions. The data allowed us to discover several differentially expressed miRNA and also novel miRNAs… BGI provides a world-class service and we will continue our project with their help, sequencing another 20 miRNA libraries”
-Dr. Marcelo Menossi, University of Campinas
Identification of MiRNomes in Human Liver and Hepatocellular Carcinoma Reveals MiR-199a/b-3p as Therapeutic Target for Hepatocellular carcinoma. Cancer Cell. 19(2):232-43 (2011).
An in-depth analysis of miRNomes in human normal liver, hepatitis liver, and hepatocellular carcinoma (HCC) was carried out in this study. We found nine miRNAs accounted for ∼88.2% of the miRNome in human liver. The third most highly expressed miR-199a/b-3p is consistently decreased in HCC, and its decrement significantly correlates with poor survival of HCC patients. Moreover, miR-199a/b-3p can target tumor-promoting PAK4 to suppress HCC growth through inhibiting PAK4/Raf/MEK/ERK pathway both in vitro and in vivo. Our study provides miRNomes of human liver and HCC and contributes to better understanding of the important deregulated miRNAs in HCC and liver diseases.
Characterization of microRNAs in Serum: a Novel Class of Biomarkers for Diagnosis of Cancer and Other Diseases. Cell Res. 18:997–1006 (2008).
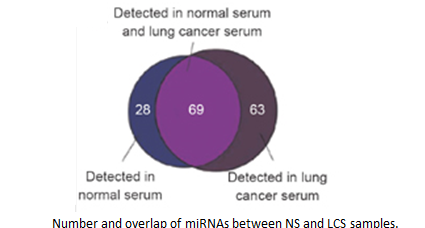 Dysregulated expression of microRNAs (miRNAs) in various tissues has been associated with a variety of diseases, including cancers. Here we demonstrate that miRNAs are present in the serum and plasma of humans and other animals such as mice, rats, bovine fetuses, calves, and horses. The levels of miRNAs in serum are stable, reproducible, and consistent among individuals of the same species. Employing Solexa, we sequenced all serum miRNAs of healthy Chinese subjects and found over 100 and 91 serum miRNAs in male and female subjects, respectively. We also identified specific expression patterns of serum miRNAs for lung cancer, colorectal cancer, and diabetes, providing evidence that serum miRNAs contain fingerprints for various diseases.
Dysregulated expression of microRNAs (miRNAs) in various tissues has been associated with a variety of diseases, including cancers. Here we demonstrate that miRNAs are present in the serum and plasma of humans and other animals such as mice, rats, bovine fetuses, calves, and horses. The levels of miRNAs in serum are stable, reproducible, and consistent among individuals of the same species. Employing Solexa, we sequenced all serum miRNAs of healthy Chinese subjects and found over 100 and 91 serum miRNAs in male and female subjects, respectively. We also identified specific expression patterns of serum miRNAs for lung cancer, colorectal cancer, and diabetes, providing evidence that serum miRNAs contain fingerprints for various diseases.
Bioinformatics Analysis:
Standard Bioinformatics Analysis
- Remove adaptors, low-quality tags, as well as contaminants to generate clean reads
- Summarize the length distribution of small RNAs
- Analyze common and specific sequences between two samples
- Explore small RNA distribution across the selected genome
- Identify rRNAs, tRNAs, snRNAs, etc. by aligning to Rfam and Genbank databases
- Identify repeat-associated small RNAs
- Identify degraded fragments of mRNAs
- Identify known miRNAs by aligning to a designated part of miRBase
- Annotate small RNAs into several categories based on priority
- Predict novel miRNAs and their secondary structures by Mireap from unannotated small RNAs
- Analyze the expression pattern of known miRNAs
- Perform family analysis of known miRNAs
Advanced Bioinformatics Analysis
- Differential expression analysis of novel and known miRNA (two or more samples should be provided) as well as cluster analysis of novel and known miRNA (three or more samples should be provided)
- Prediction of novel and known miRNA target genes (only for one single sample)
- Gene Ontology (GO) annotation and KEGG pathway analysis of novel and known miRNA target genes (only for one single sample)
- Base editing analysis of miRNA
- Prediction of differential miRNA target genes(two or more samples should be provided)
- GO annotation and KEGG pathway analysis of differential miRNA target genes (two or more samples should be provided)
Custom Bioinformatics Analysis
- We can also perform other customized analyses to meet the requirements of specific projects.
Sample Requirements:
- Sample condition: Integrated total RNA samples. Avoid protein contamination during RNA isolation.
- Sample quantity (for every single library construction):
- General requirements: total RNA ≥ 10 μg (standard) , total RNA≥200 ng (minimum)
- For plasma/serum samples: total RNA ≥100 ng
- For miRNA (<200nt): RNA ≥1 μg
- Sample concentration:
General requirements: concentration ≥200ng/μL
For plasma/serum samples: concentration ≥ 2ng/μL
For miRNA (<200nt): concentration ≥20 ng/μL - Sample purity (for eukaryotes except insects): 28S:18S ≥1.5, RIN ≥8.0
Turnaround Time:
The standard turnaround time for the workflow (above) is 30 business days.
Completion Indicator:
The completion is indicated by the number of the clean reads. Goals are individualized for each project.

