- Genomics
- Transcriptomics
- Epigenomics
- Meta-omics
- Proteomics
- Single-Cell Sequencing
- Immune Repertoire Sequencing
- FFPE Samples

ALLinONE
Cases
Technical Information
Contact Us / Wish List
ALLinONE, developed by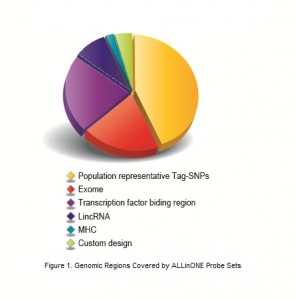 BGI, is an innovative and integrated human genome capture array for next generation sequencing. ALLinONE captures 5-10% of the human genome including whole exome, population representative tagSNP region, MHC region as well as other customized target region, and is a cost-effective method for whole genome screening by providing more information than exome sequencing. ALLinONE utilizes ethnicity-specific sequence capture solution, and combines different advantages of exome capture sequencing, genotyping chips and custom-designed chips all together. ALLinONE is expected to become a powerful new-generation tool for human disease genomics, phenotype-genotype association researches, and other human population genetic studies.
BGI, is an innovative and integrated human genome capture array for next generation sequencing. ALLinONE captures 5-10% of the human genome including whole exome, population representative tagSNP region, MHC region as well as other customized target region, and is a cost-effective method for whole genome screening by providing more information than exome sequencing. ALLinONE utilizes ethnicity-specific sequence capture solution, and combines different advantages of exome capture sequencing, genotyping chips and custom-designed chips all together. ALLinONE is expected to become a powerful new-generation tool for human disease genomics, phenotype-genotype association researches, and other human population genetic studies.
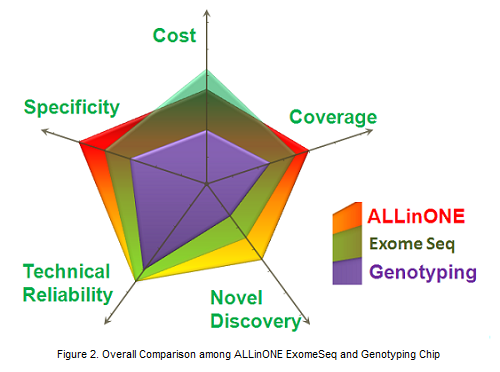
Benefits:
Population Specificity: ALLinONE is designed towards specific population, which will greatly increase the power of genetic association study in epidemiology.
Data Comprehensiveness: The entire genomic coding region, most of population haplotype blocks and MHC region are covered.
Data Flexibility: Custom-designed probes can be added according to particular research interests.
Technical Reliability: High coverage of target region and data accuracy.
High Efficiency: Highly cost effective, both common and rare variants will be detected.
Customer Testimonials:
“The ALLinOne capture technique will be the "golden middle road" between whole-genome sequencing, which is still cost prohibitive for thousands of samples, and exome sequencing, which does not capture enough of the genome” – Prof. Lennart Hammarstöm, Karolinska Institute University Hospital.
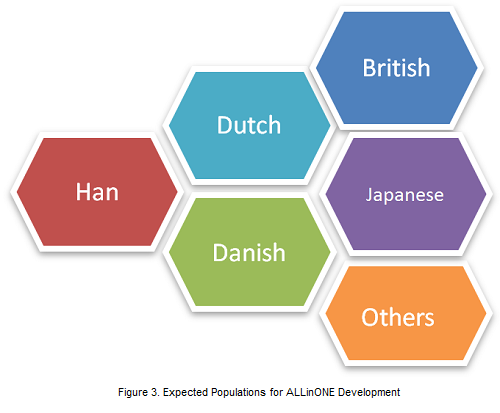
BGI has already developed two sorts of ALLinONE arrays for Han Chinese. Based on different tagSNP number, the arrays are named Han ALLinONE (1.3M) and Han ALLinONE (690K). Han ALLinONE (1.3M) has already been used in disease genomic studies. In the near future, we plan to develop ALLinONE arrays for Dutch, Danish, British, Japanese, and other populations. In addition, we expect to find more collaborators who are interested in this solution.
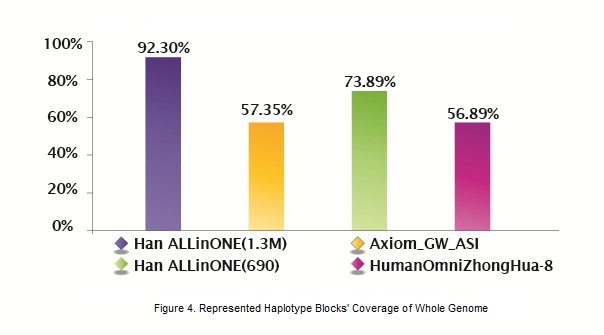
 To assess the validity of the tagSNPs selected, we compared ALLinONE with two genotyping products, Axiom developed by Affymetrix and HumanOmniZhongHua-8 developed by Illumina. The figure below compares the coverage of the whole genome represented by the tagSNP region and demonstrates that AllinOne provides much higher coverage.
To assess the validity of the tagSNPs selected, we compared ALLinONE with two genotyping products, Axiom developed by Affymetrix and HumanOmniZhongHua-8 developed by Illumina. The figure below compares the coverage of the whole genome represented by the tagSNP region and demonstrates that AllinOne provides much higher coverage.
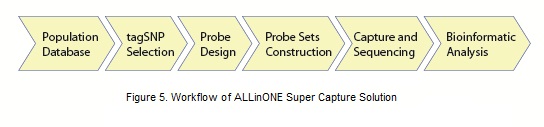
To develop ALLinONE arrays for specific populations, a reliable population database is used (e.g., 1000 Genomes Project). If no database is available, whole genome sequencing of at least 100 samples is recommended to construct an effectual database for the population. TagSNPs are selected based on the linkage disequilibrium information in the population database, and probes are designed to cover these tagSNPs. The whole probe set is included in ALLinONE arrays to perform target region capture and sequencing.
Sample Requirements:
For the genomic DNA samples provide the following:
- Purity: OD260/280 = 1.8-2.0, without degradation and RNA contamination
- Concentration: ≥ 50ng/μl
- DNA amount: single library preparation starts from at least 6μg, the more the better

