
- Genomics
- Transcriptomics
- Epigenomics
- Meta-omics
- Proteomics
- Single-Cell Sequencing
- Immune Repertoire Sequencing
- FFPE Samples

BAC/Fosmid End Sequencing
End sequencing is used primarily for choosing purpose clones and estimating the results of the genome assembly.
Workflow:
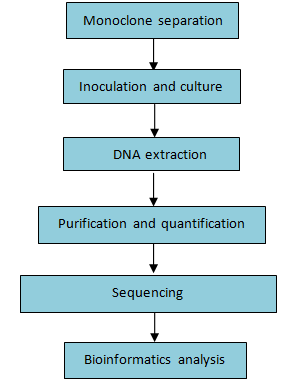 Figure 7. Workflow of BAC/Fosmid end sequencing
Figure 7. Workflow of BAC/Fosmid end sequencing
Bioinformatics Contents:
Bioinformatics content consists of basic data analysis processing, including base calling, vector removal, etc.
Sample Requirements:
For fresh bacteria provide a single sample size of at least 500 μL or a batch sample of at least 50 μL in a 96-well plate; Vector information, sequencing primer, resistance, and fragment size should be confirmed.
Turnaround Time:
For inducible samples the standard turnaround time using the workflow (above) is 6 to 7 working days.
Completion Milestone:
The project completion milestone is based on a quality score of(Q20), with read lengths of > 450 bp.
