
Bioinformatics Analysis
With the rapid development of sequencing technology, higher throughput of better quality data can now be achieved at a lower cost. These data are being extensively used to decipher the mechanisms of biological systems at the most basic level. However, this huge and growing amount of high-quality data and the information delivered continue to create challenges for bioinformaticians.
There are several bioinformatics sub-groups at BGI focusing on analyzing and transferring cutting-edge technologies in their respective fields to maximize the amount of information that can be gleaned from sequencing data. Their analyses include:
 Data Processing and Quality Control
Data Processing and Quality Control
Assembly
Mapping
Annotation
Advanced Analysis
Data Processing and Quality Control
Data processing consists of three steps: image analysis, base calling and sequence analysis. QC is an important and effective measure for determining sample libraries’ qualities, and it also serves to indicate whether the sequencing succeeded or failed. Read sequences are aligned by employing BWA (Burrows-Wheeler Aligner) and data are produced in BAM format to conduct further analysis.
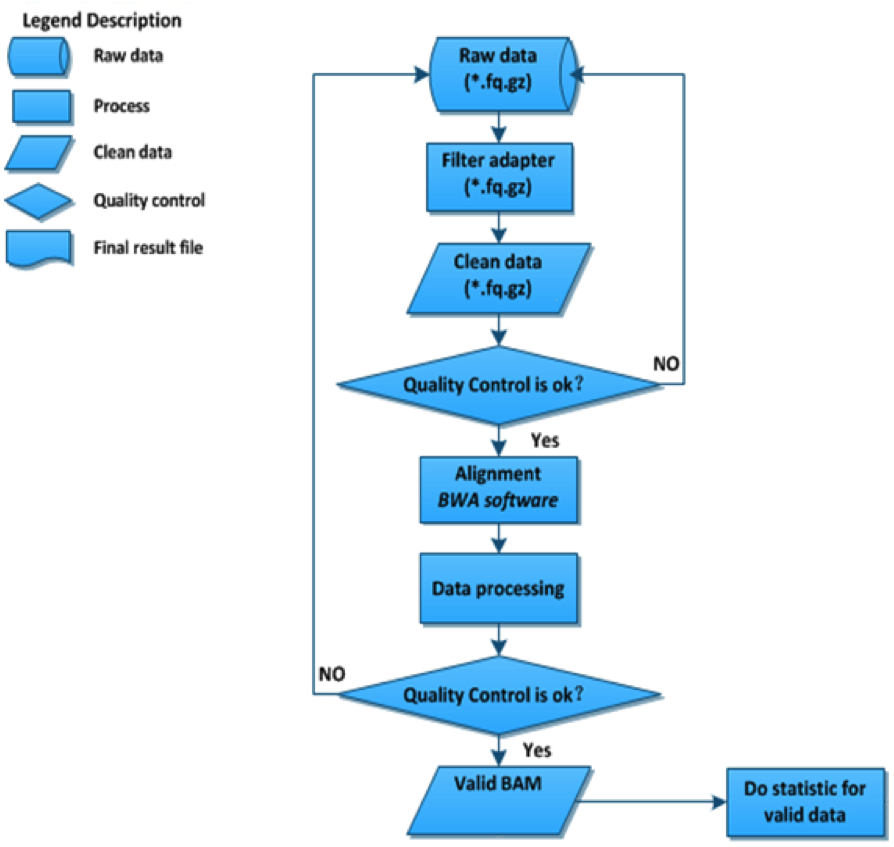
The workflow for NGS data processing and QC in BGI.
Assembly
Assembly involves methods to assemble short-sequencing reads into entire and accurate reference genomes. The primary work of this division is to develop and improve assembly strategies, especially using short-read sequencing technologies.
Genome de novo assembly
SOAPdenovo, developed by BGI, is an assembler that has successfully carried out de novo assembly of several large genomes, including the cucumber, Asian individuals, the Giant Panda, and others.
Transcriptome de novo assembly
Transcriptome de novo assembly is carried out using SOAPdenovo. Sequences obtained from multiple samples of the same species are sequenced, and then unigenes from each sample assembly are further processed with our sequence clustering software to remove sequence splicing and redundancy to acquire a non-redundant unigene set.
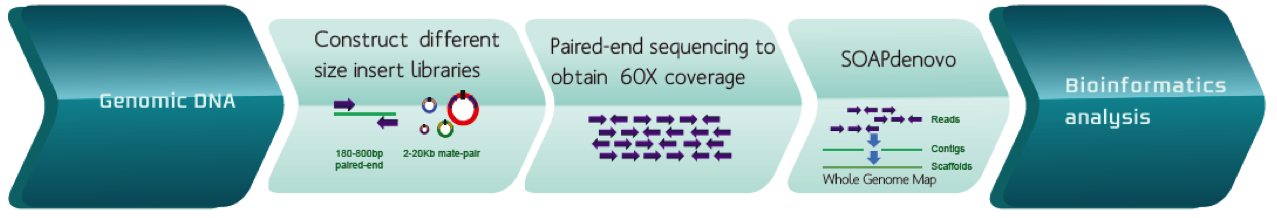
An example of de novo assembly workflow for genomic DNA.
Mapping
Mapping involves methods used to compare novel sequences with a reference genome to detect variations, and to analyze different expression levels.
Genome mapping
For whole genome mapping, we use SOAPaligner software with suitable parameters to map the whole genome adapter-free reads to the reference genome. Mutations like SNP, SV, SNV, CNV, and InDel are detected in addition to the methylation status of cytosine in single-base-pair resolution for bisulfite-pretreated DNA samples.
For target region mapping, we use SOAPaligner software to map reads to a reference genome as well, but use the reads that are sequenced from DNA captured by enzymes, antibodies, or designed chips. Mutations like SNPs in the exome regions (especially in CDS region), and short InDels are detected, followed by further studies on novel and functional mutations. For MeDIP and Chip-Seq, positions of methylated DNA regions can be determined by the target region mapping.
Transcriptome mapping using reference data
To carry out transcriptome mapping, low quality reads and reads that contain adaptor sequences are filtered out from the raw data. SOAPaligner software, developed at BGI, is then used to map these data onto a reference genome and genes. Only two mismatches are allowed in these alignments. Novel transcripts are then identified in the intergenic region of the genome using transcriptional activity as evidence. Gene expression levels are determined by the number of reads that map to each gene and presented in Reads per kb per Million mapped reads (RPKM).
Small RNA mapping
For small RNA mapping, we first generate a set of clean reads by removing low quality tags and other contaminants, such as tags without 3’ adaptors and tags with 5’ adaptors. The length distribution of these clean reads is summarized to provide information on the small RNA composition of each sample. Clean reads are then mapped—allowing no mismatches—onto a reference genome using the BGI designed SOAPaligner software to locate each read on the genome sequences.
Annotation
Annotation includes methods to add biological information to raw DNA sequence, identify the structural and functional elements, and integrate and display this information at a genomic level.
Genome annotation
Genome annotation is the process of adding biological information to raw DNA sequence that has been produced in genome-sequencing projects. The value of the genome is only as good as its annotation. Therefore, it is necessary to obtain the highest quality annotation for each genome. The goal of annotation is to identify the key features of the genome, in particular protein-coding genes and their products. In addition to protein-coding genes, it is also possible to identify repeats, non-coding RNAs, and some regulatory elements using de novo or homology-based methods.
Transcriptome annotation
To annotate the transcriptome, we perform Blast alignment (e-value < 0.00001) between the Unigene set and protein databases, including nr, Swiss-Prot, KEGG, and COG. The results from the best alignment are used to determine the direction of Unigenes. In cases where the results from different databases conflict, we use the following order of priority: nr, Swiss-Prot, KEGG, and COG. Our functional annotation process of the Unigenes provides protein functional annotation, COG functional annotation, and Gene Ontology (GO) functional annotation.
Small RNA annotation
We align and annotate clean reads with ncRNA in Genbank and Rfam, repeat-associated RNA, exons and introns in mRNAs, and miRNAs each into one category based on priority. All known miRNA families identified in a sample are investigated for their presence in other species. As there might be mutations in miRNAs in the sample, known miRNA from a reference are used to identify these mutations by comparing the newly sequenced miRNA to the known miRNA. The remaining reads that cannot be annotated into any category, are considered potential candidates for novel miRNAs and are assessed using Mireap. For novel miRNA candidates, we predict their targets based on their complementary sequences and the quality of their secondary structure.
Advanced Analysis
BGI also provides advanced bioinformatics analysis for various research purposes, such as Mendelian diseases, complex diseases, cancer and population analysis.
Mendelian Diseases
Mendelian disorder refers to human disorder caused by mutation in a single gene. To find disease-causing mutations, we conduct diverse bioinformatics analyses. First, detected SNPs and Indels are filtered with public databases including dbSNP, the 1000 Genomes project, eight HapMap exomes and the first Asian genome. Second, SV, CNV, rare SNP and Indel shared by all cases are obtained and then filtered with control if available. In addition, amino acid substitution prediction with SIFT, and annotation with KEGG and GO databases can be done for candidate mutation genes. HMM prediction can also be done for pedigree with two or more cases.
Complex Diseases
With the aim to find disease-associated variants, the advanced analysis of PLINK, PCA etc, are available from BGI for large scale samples or reasonably selected samples but with smaller number of variants. Moreover, for family-base samples, the advanced analysis of de novo mutation detection is available as well.
Cancer
Cancer bioinformatics analyses focus on somatic mutations, including SNP/InDel/SV, which occur in somatic cells after conception (except for familial cancer). For paired normal-tumor samples, tumor specific mutations (SNV/CNV) can be detected and annotated precisely. In addition, other structural variants, like rearrangements and virus integration deletions, can also be detected. Functional analysis, like amino acid substitution prediction, pathway analysis, and GO enrichment analysis, can be done for mutation genes that may have a functional relationship with carcinogenesis. Mutation target network analysis, selection pressure analysis and driver gene detection are appropriate for studies with larger sample quantities.
Population Analysis
Each human population is different from others due to geographic distribution, environment and other exogenous factors. To find out these population-specific features and infer human evolution history, we conduct diverse bioinformatics analyses which include ancestral analysis, population structure analysis and selection signal analysis based on a huge amount of variants from population-scale sequencing data. Meanwhile, we will construct a haplotype map for different populations since every population has its own LD pattern and haplotype structure and frequencies.
Comparative Genomics
Comparative Genomics includes:
- Identification of orthologous and paralogous genes from several species.
- Estimation of the divergence time between species, the average evolutionary rate, and selective pressure.
- Analysis of evolution in the size of gene families.
- Detection of positively selected genes and genomic regions.
- Identification of evolutionary relationships and chromosomal rearrangement between genomes.
- Detection of segmental duplication and whole-genome duplication events.
- Analysis of genes or genomic regions that are related to species-specific biological characteristics.

