- Genomics
- Transcriptomics
- Epigenomics
- Meta-omics
- Proteomics
- Single-Cell Sequencing
- Immune Repertoire Sequencing
- FFPE Samples

Human MHC-Seq
Cases
Technical Information
Contact Us / Wish List
 The major histocompatibility complex (MHC) plays a critical role in hundreds of diseases and is a key target in drug research and development. In humans, it is also called human leucocyte antigen (HLA) and lies on the short arm of chromosome 6. BGI and Roche NimbleGen have developed an MHC region capture technology (MHC-Seq) to allow easy capture, enrichment, and deep sequencing of these highly repetitive regions. MHC-Seq targets the traditional MHC region of approximately 3.37 Mb (HG19: chr6: 29691116-33054976) as well as 1.6 Mb surrounding the MHC region (a total of 4.97 Mb), and includes eight known haplotypes (PGF, APD, COX, DBB, MANN, MCF, QBL, and SSTO). This new technology can type 26 HLA genes with 4-digit resolution and detect variants such as SNPs and InDels.
The major histocompatibility complex (MHC) plays a critical role in hundreds of diseases and is a key target in drug research and development. In humans, it is also called human leucocyte antigen (HLA) and lies on the short arm of chromosome 6. BGI and Roche NimbleGen have developed an MHC region capture technology (MHC-Seq) to allow easy capture, enrichment, and deep sequencing of these highly repetitive regions. MHC-Seq targets the traditional MHC region of approximately 3.37 Mb (HG19: chr6: 29691116-33054976) as well as 1.6 Mb surrounding the MHC region (a total of 4.97 Mb), and includes eight known haplotypes (PGF, APD, COX, DBB, MANN, MCF, QBL, and SSTO). This new technology can type 26 HLA genes with 4-digit resolution and detect variants such as SNPs and InDels.
Benefits:
- Co-developed by BGI and NimbleGen
- The first kit to cover the entire MHC region (a total of 4.97 Mb)
- High coverage: > 97% of MHC and extended regions
- High accuracy: > 95% of 26 HLA genes can be typed at 4-digit resolution
- The best choice for immunology research with lower costs
Applications:
- Investigation of variations associated with HLA-immune diseases
- Vaccine and drug target screening
- Organ transplant matching
- Paternity testing and forensic science
An Integrated Tool to Study MHC Region: Accurate SNV Detection and HLA Genes Typing in Human MHC Region Using Targeted High-Throughput Sequencing. Plos one. DOI: 10.1371/journal.pone.0069388 (2013).
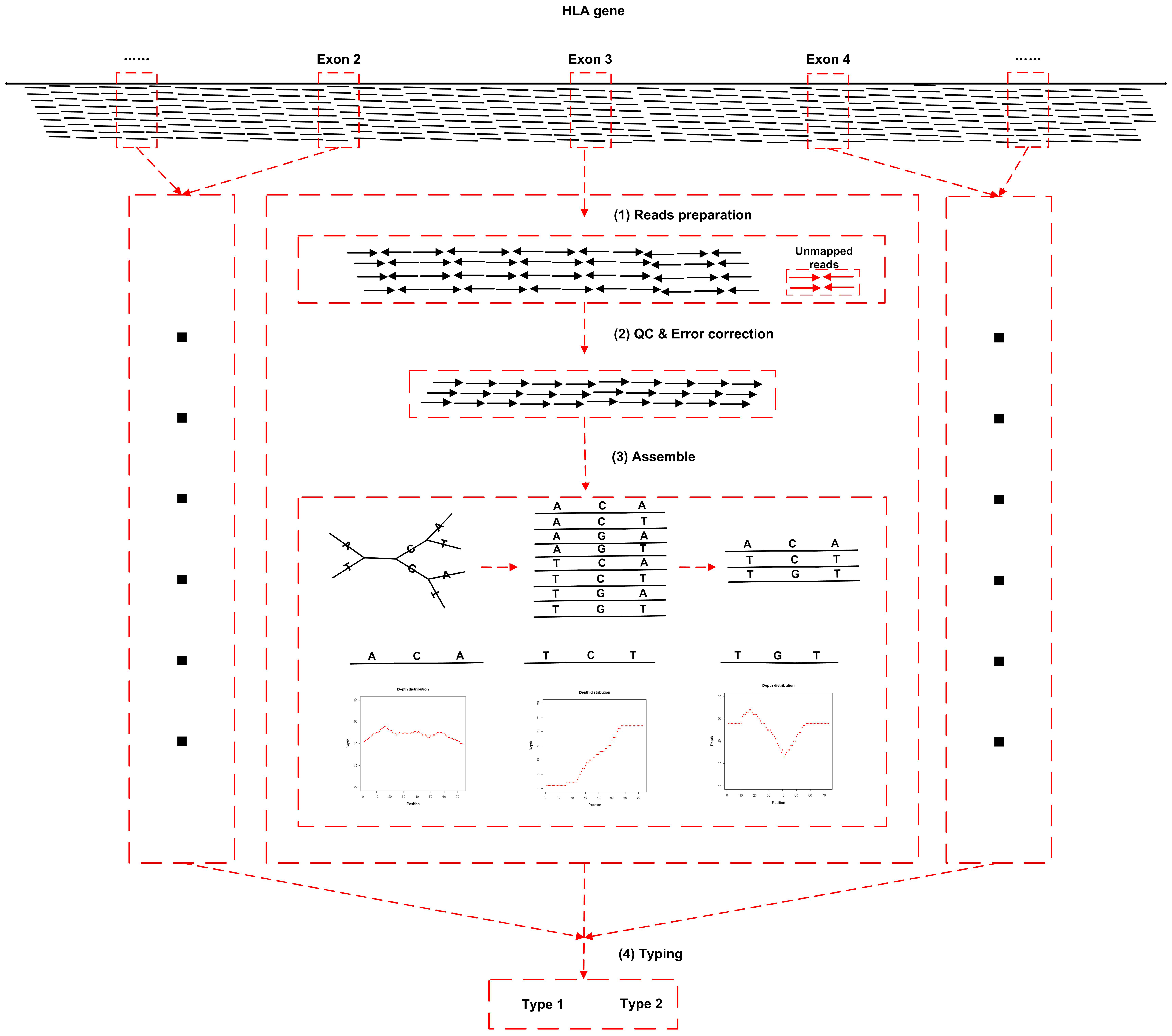 The study presented a method for the accurate detection of minor variants and HLA types in the human MHC region using high-throughput, high-coverage sequencing of target regions. A probe set was designed as a template based on eight annotated human MHC haplotypes and it encompasses 5 megabases (Mb) of the extended MHC region. The results showed that >97% of the overall MHC region and >99% of the gene coding regions were covered with sufficient depth and good evenness. Moreover, 98% of genotypes called by this capture sequencing were proved to be consistent with established HapMap genotypes. In addition, we have concurrently developed a one-step pipeline (the left figure) for calling any HLA type referenced in the IMGT/HLA database from this target capture sequencing data has been developed, which shows over 96% typing accuracy when deployed at 4 digital resolution.
The study presented a method for the accurate detection of minor variants and HLA types in the human MHC region using high-throughput, high-coverage sequencing of target regions. A probe set was designed as a template based on eight annotated human MHC haplotypes and it encompasses 5 megabases (Mb) of the extended MHC region. The results showed that >97% of the overall MHC region and >99% of the gene coding regions were covered with sufficient depth and good evenness. Moreover, 98% of genotypes called by this capture sequencing were proved to be consistent with established HapMap genotypes. In addition, we have concurrently developed a one-step pipeline (the left figure) for calling any HLA type referenced in the IMGT/HLA database from this target capture sequencing data has been developed, which shows over 96% typing accuracy when deployed at 4 digital resolution.
Workflow
The workflow of MHC capture sequencing is similar to target region sequencing. Genomic DNA is tested for quality then subjected to library construction with an insert size of 500 bp, followed by PE 91 sequencing on Illumina HiSeq 2000 platforms. The recommended sequencing depth is at least 80X. For bioinformatics analysis, basic variants such as SNPs and InDels will be called, and 26 HLA genes can be typed. The 26 genes include A, B, C, DRB1, DQB1, DPA1, DPB1, DQA1, G, DMA, DMB, DOA, DOB, DRA, E, F, H, J, K, MICA, MICB, P, TAP1, TAP2, V ,and L.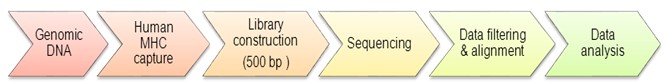
Bioinformatics
Standard Bioinformatics Analysis
- Data filtering (removing contaminating adapters and low-quality reads from raw reads)
- Alignment and summary of data
- Duplicates removal
- Local realignment around InDels and base quality re-calibration
- SNP calling, annotation, and statistics
- InDel calling, annotation, and statistics
- HLA typing of 26 genes at 4-digit resolution
Advanced Bioinformatics Analysis
- Association analysis between diseases and mutations (SNPs and InDels)
- Association analysis between diseases and HLA types (26 genes)
Sample Requirements
For genomic DNA samples please provide:
- Purity: OD260/280 = 1.8 - 2.0, without degradation or RNA contamination
- Concentration: ≥ 37.5 ng/μL
- Quantity: single library preparation starts from at least 1 μg (although ≥2.5 μg is recommended)
Turnaround Time
The standard turnaround time for 100 samples with standard workflow (80x) is about 50 working days (kit ordering is not included) (Table 1).Table 1. Turnaround Time for MHC Capture Sequencing
| Sample size | Kit Ordering (days) | Library Construction & PE 91 Sequencing (days) | Bioinformatics Analysis (days) |
| X≤48 | 60 | 37 | 10 |
| 48≤X≤144 | 60 | 44 | 10 |
| 144≤X≤288 | 60 | 51 | 10 |

