
Exome & Target Region Sequencing
Cases
Technical Information
Contact Us / Wish List
 Exome sequencing selectively targets the most functionally relevant DNA sequences that encode proteins, allowing the identification of novel genes associated with both Mendelian disorders and common diseases. Target region capture enriches specific regions or genes of interest by microarray (NimbleGen Sequence Capture Array) or solution hybridization (Agilent Sure-Select™ system) methods.
Exome sequencing selectively targets the most functionally relevant DNA sequences that encode proteins, allowing the identification of novel genes associated with both Mendelian disorders and common diseases. Target region capture enriches specific regions or genes of interest by microarray (NimbleGen Sequence Capture Array) or solution hybridization (Agilent Sure-Select™ system) methods.
BGI is highly experienced in exome sequencing and analysis for human, plants, and animals. To date, we have sequenced more than 38,000 human exomes. In addition, we provide mouse exome sequencing service using Agilent Mouse All Exon kit and developed the exome sequencing platform for monkeys based on our research from the Chinese rhesus macaque and Cynomolgus macaque genome projects.
Click here to learn more about our latest promotion for human exome sequencing (50X, 100X) valid through May 31, 2013.
Benefits:
- Deep experience in exome sequencing
- Rapid turnaround at our Hong Kong and CHOP (Philadelphia, PA) facilities
- High quality data (See examples of data generated by BGI)
- Affordable pricing
- Strong analytical capabilities by 1,000 bioinformaticians (Advanced analysis available for Mendelian disorder, complex disease, cancer, population analysis)
Customer Testimonial:
"We use BGI as a NGS service provider for many of our targeted region re-sequencing projects. BGI has consistently provided high-quality data with a very quick turnaround time. I know exactly what to expect in terms of delivery time and data quality, which is vital for our business operations."
-Dr. Eric Lin, Chairman of the Board, Otogenetics Corporation
BGI has successfully completed numerous exome sequencing projects, including a Danish study of 1000 patient samples and 1000 controls with the aim of finding rare SNPs associated with metabolic disorders such as obesity and hypertension.
Frequent Mutations of Genes Encoding Ubiquitin-mediated Proteolysis Pathway Components in Clear Cell Renal Cell Carcinoma. Nature Genetics. 44:17-9 (2012).
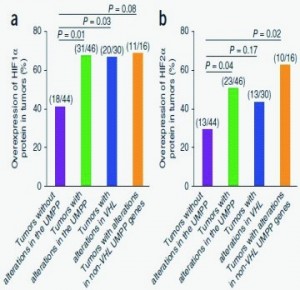 The research sequenced whole exomes of ten clear cell renal cell carcinomas (ccRCCs) andperformed a screen of ~1,100 genes in 88 additional ccRCCs. Frequent mutations weredetected in the ubiquitin-mediated proteolysis pathway (UMPP). The findings highlight thepotential contribution of UMPP to ccRCC tumorigenesis through the activation of thehypoxia regulatory network.
The research sequenced whole exomes of ten clear cell renal cell carcinomas (ccRCCs) andperformed a screen of ~1,100 genes in 88 additional ccRCCs. Frequent mutations weredetected in the ubiquitin-mediated proteolysis pathway (UMPP). The findings highlight thepotential contribution of UMPP to ccRCC tumorigenesis through the activation of thehypoxia regulatory network.
Exome Sequencing Identifies NMNAT1 Mutations as a Cause of Leber Congenital Amaurosis. Nature Genetics. 44:972-4 (2012).
 The exome of an individual with Leber congenital amaurosis£¨LCA£©was sequenced and identifiednonsense (c.507G>A, p.Trp169*) and missense (c.769G>A, p.Glu257Lys) mutations in NMNAT1,which encodes an enzyme in the nicotinamide adenine dinucleotide (NAD) biosynthesis pathway.It is implicated in protection against axonal degeneration. We also found NMNAT1 mutations inten other individuals with LCA, all of whom carry the p.Glu257Lys variant.
The exome of an individual with Leber congenital amaurosis£¨LCA£©was sequenced and identifiednonsense (c.507G>A, p.Trp169*) and missense (c.769G>A, p.Glu257Lys) mutations in NMNAT1,which encodes an enzyme in the nicotinamide adenine dinucleotide (NAD) biosynthesis pathway.It is implicated in protection against axonal degeneration. We also found NMNAT1 mutations inten other individuals with LCA, all of whom carry the p.Glu257Lys variant.
Single-Cell Exome Sequencing Reveals Single-Nucleotide Mutation Characteristics of a Kidney Tumor. Cell. 148:886-95 (2012).
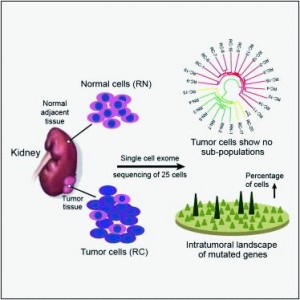 To better understand the intratumoral genetics underlying mutations of ccRCC, single-cellexome sequencing was carried out on a clear cell renal cell carcinoma (ccRCC) and its adjacentkidney tissue. The pilot study demonstrates that ccRCC may be more genetically complex thanpreviously thought and provides information that can lead to new ways to investigateindividual tumors, with the goal of developing more effective cellular targeted therapies.
To better understand the intratumoral genetics underlying mutations of ccRCC, single-cellexome sequencing was carried out on a clear cell renal cell carcinoma (ccRCC) and its adjacentkidney tissue. The pilot study demonstrates that ccRCC may be more genetically complex thanpreviously thought and provides information that can lead to new ways to investigateindividual tumors, with the goal of developing more effective cellular targeted therapies.
An Integrated Map of Genetic Variation from 1,092 Human Genomes. Nature. 491:56-65 (2012).
 By characterizing the geographic and functional spectrum of human genetic variation, the 1000Genomes Project aims to build a repository that can help understanding the geneticcontribution to disease. Up to 98% of accessible single nucleotide polymorphisms at a frequencyof 1% in related populations were captured, enabling analysis of common and low-frequency variants in individuals from diverse, including admixed, populations.
By characterizing the geographic and functional spectrum of human genetic variation, the 1000Genomes Project aims to build a repository that can help understanding the geneticcontribution to disease. Up to 98% of accessible single nucleotide polymorphisms at a frequencyof 1% in related populations were captured, enabling analysis of common and low-frequency variants in individuals from diverse, including admixed, populations.
Sequencing of 50 Human Exomes Reveals Adaptation to High Altitude.
Science. 329, 75 (2010).

50 exomes of ethnic Tibetans were sequenced for 18X per individual. Genes showing population-specific allele frequency changes, which represent strong candidates for altitude adaptation, were identified. One single-nucleotide polymorphism (SNP) at EPAS1 shows a 78% frequency difference between Tibetan and Han samples, representing the fastest allele frequency change observed at any human gene to date. This research can help us to prevent and cure the disease of plateau anoxia.
Bioinformatics:
Standard Bioinformatics Analysis
- Data filtering (removing adaptors contamination and low-quality reads from raw reads)
- Alignment and summary of data production
- SNP calling, InDel calling, annotation and statistics
Population Advanced Analysis
- InDel calling, annotation and statistics
- Population SNP calling
- Population InDel calling
- Haploview:
- Linkage disequilibrium
- Haplotype prediction
- Positive selection signals detection
Cancer Advanced Analysis
- Somatic SNP/InDel detection for paired normal-tumor samples
- SNV detection, annotation and statistics for paired normal-tumor samples
- Filtering with the known databases like cosmic, dbSNP etc.
- Amino acid substitution prediction (SIFT, Polyphon-2)
- GO enrichment analysis for selected genes and pathway enrichment analysis
Complex Disease Advanced Analysis
- InDel calling, annotation and statistics
- NGS-GWAS
- Population SNP calling, minor allele frequency (MAF) estimation and association analysis for low depth and large sample amount
- PLINK-based association analysis for deep sequenced large sample scale or reasonable selected fewer samples
- De novo mutation detection for family-based samples
Mendelian disorders Advanced Analysis
- Gender judgement
- InDels calling, annotation and statistics
- Screening for mutations in coding regions, UTR and splicesite regions
- Filtering with databases
Exome Capture Arrays:
BGI currently has the capacity to process 800 samples of exome capture per week. There are mainly two exome capture strategies which both deliver a high-level performance and substantial savings on sequencing. Both NimbleGen SeqCap EZ (Biotinylated DNA oligonucleotide probes) and Agilent SureSelect system (Biotinylated RNA probes) can capture all exons in solution via a simple, scalable workflow and stringent built-in quality controls.
Exome capture arrays that we perform are as follows:
Human Exon Capture Array:
| Design | Capture Targets (Mb) (Regions Covered by Probes) | Database Used to Select Primary Targets |
|
Agilent SureSelect Human All V3 |
50 |
CCDS Sep 2009 + miRBase V14 + GENCODE + Sanger |
|
Agilent SureSelect Human All V4 |
51 |
CCDS Mar 2011 + miRBase V17 + GENCODE + RefSeq Mar 2011 |
|
Agilent SureSelect Human All V4+UTRs |
71 |
CCDS Mar 2011 + miRBase V17 + GENCODE + RefSeq Mar 2011 |
|
NimbleGen SeqCap EZ Exome V2.0 |
44 |
CCDS Sep 2009 + miRBase V14, Sep 2009 + RefSeq Jan 2010 |
|
NimbleGen SeqCap EZ Exome V3.0 |
64 |
CCDS Apr 2011 + miRBase V15 + GECODE+RefSeq Jun 2011 |
Mouse Exome Capture Array:
| Exome capture kit | Insert size | |
|
Agilent SureSelect Mouse |
450 Mb |
150-200 bp |
|
NimbleGen Maize All |
36 Mb |
200-300 bp |
Target Region Sequencing:
| Main capture arrays or kits | Personalized capture kits |
|
NimbleGen human 2.1M custom array |
Agilent SureSelectXT Human Kinome Panel Kit |
|
Agilent Sureselect XT Custom Kit |
NimbleGen Human HLA kit |
|
NimbleGen SeqCap EZ Choice XL library |
Human X-Chromosome exon Kit |
Sample Requirements:
Human Exome Sequencing:
For the genomic DNA samples you will provide us:
- Purity: OD260/280=1.8~2.0, without degradation and RNA contamination
- Sample concentration: ≥30ng/μl
- Quantity demanded: 1μg (3μg gDNA recommended)
Human Target Region Sequencing:
For the genomic DNA samples you will provide us:
- Purity:OD260/280=1.8-2.0, without degradation and RNA contamination
- Concentration: ≥30ng/μl
- Quantity demanded: ≥30μg (using NimbleGen Sequence Capture Array when the targeted region is larger than 17Mb) or ≥6μg (using Agilent SureSelect System/NimbleGen Sequence Capture Array when the targeted region is smaller than 17Mb).
Mouse Exome/Target Region Sequencing:
For the genomic DNA samples you will provide us:
- Purity: OD 260/280=1.8~2.0, without degradation and RNA contamination
- Concentration: ≥30ng/μl
- Quantity demanded: ≥30μg (using NimbleGen Sequence Capture Array when the targeted region is larger than 17Mb) or ≥6μg (using Agilent SureSelect System or using NimbleGen Sequence Capture Array when the targeted region is smaller than 17Mb).
Turnaround Time:
The standard turnaround time for the workflow (above, exclude advanced informatics analysis) is about ~40 working days for exome sequencing of 100 samples with 50X coverage;
The standard turnaround time for the workflow (As figure above: exclude advanced informatics analysis) is ~140 working days (including chip customization and pilot study) for 1000 samples with about 1Mb target region size.

