
Metagenomics
Cases
Technical Information
Contact Us / Wish List
 Metagenomics is the study of genomes of a whole microbial community. It has opened up a new era in the study of microbial diversity with direct access to the genomes of numerous uncultivable microorganisms in their natural habitat. Metagenomics has been applied in environmental studies as well as biomarker research. BGI offers a complete range of metagenomics solutions:
Metagenomics is the study of genomes of a whole microbial community. It has opened up a new era in the study of microbial diversity with direct access to the genomes of numerous uncultivable microorganisms in their natural habitat. Metagenomics has been applied in environmental studies as well as biomarker research. BGI offers a complete range of metagenomics solutions:
- 16S rDNA tagging: general taxonomics classification
- Whole genome metagenomic sequencing
- Establish microbial gene catalogue
- Human microbe & related disease association analysis
- Metatranscriptome survey
Benefits:
- No need to culture samples (Most microbes non-culturable)
- Study microbes in the native environment
- Detect more novel microbes and genes from the environmental microbial community
- High-throughput, cost-effective reads and sequencing
- More accurate data due to lack of sample bias
Customer Testimonials:
"BGI has demonstrated incredible enthusiasm and resources in terms of sequencing capacity, technical development, and informatics expertise. The EMP is the largest sequencing project ever undertaken, and BGI is the biggest player in global sequencing so their participation is essential. BGI's commitment to scientific excellence makes them an ideal choice for developing a project of this scale." Jack A. Gilbert, Ph.D., project director of Earth Microbiome Project and professor of University of Chicago and Argonne National Laboratory
A Human Gut Microbial Gene Catalogue Established by Metagenomic Sequencing. Nature. 2010; 464:59-65
 In 2010, BGI established the first human microbial gene catalogue by Metagenomics sequencing. In order to understand the impact of gut microbes on human health and well-being, 124 faecal samples obtained from European individuals were sequenced. This study confirmed that bacterial species abundance and bacterial genes differentiated between healthy individuals and disease affected patients. These findings offer an important theoretical basis for further exploring the relationship between human gut microbes, obesity, enteritis, and other diseases.
In 2010, BGI established the first human microbial gene catalogue by Metagenomics sequencing. In order to understand the impact of gut microbes on human health and well-being, 124 faecal samples obtained from European individuals were sequenced. This study confirmed that bacterial species abundance and bacterial genes differentiated between healthy individuals and disease affected patients. These findings offer an important theoretical basis for further exploring the relationship between human gut microbes, obesity, enteritis, and other diseases.
Workflow:
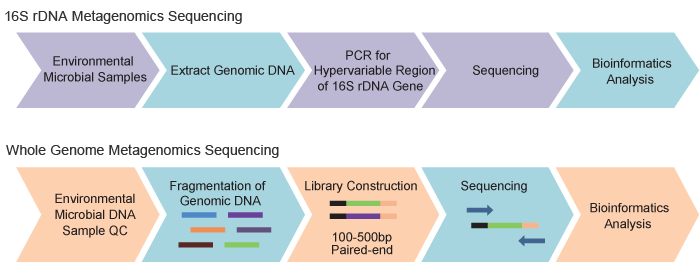 Figure 1. BGI Metagenomics Sequencing Workflow
Figure 1. BGI Metagenomics Sequencing Workflow
Bioinformatics Analysis:
- Species composition analysis
- Short reads assembly
- Gene prediction
- Gene functional annotation
- Statistical analysis of the results.
For multiple samples, the comparative analysis between different samples and the association studies can also be provided.
Sample Requirements:
Metagenomics sequencing should be discussed case by case, and a proposal can be provided after the discussion of your project requirements.
Whole Genome Metagenomic Sequencing/ Establishing a Microbial Gene Catalogue:
For the microbial genomic DNA you provide us:
- Purity:OD260/280=1.8-2.0
- Concentration: ≥50ng/μl
- DNA amount: single library preparation starts from at least 5μg, and the total amount should be determined case by case.
16S rDNA Tagging:
For the PCR products samples you provides us:
- Purity: OD260/280:1.8-2.0
- Concentration: ≥50ng/μl
- DNA amount: single library preparation starts from at least 5μg, and the total amount should be determined case by case.

