
RNA-Seq (Quantification)
Cases
Technical Information
Contact Us / Wish List
 RNA-Seq (Quantification) is used to analyze gene expression of certain biological objects under specific conditions. It is a cost-effective quantification method that produces greater sensitivity, reproducibility, and wider dynamic range than microarrays. BGI quantifies whole-genome gene expressions using next-gen sequencer Illumina HiSeq 2000 and provides differential gene expression and pathway analysis. Powered by our unique bioinformatics capabilities, BGI’s RNA-Seq services deliver reliable data and analyses to our customers conducting drug response studies, biomarker detection, basic medical research, and drug R&D.
RNA-Seq (Quantification) is used to analyze gene expression of certain biological objects under specific conditions. It is a cost-effective quantification method that produces greater sensitivity, reproducibility, and wider dynamic range than microarrays. BGI quantifies whole-genome gene expressions using next-gen sequencer Illumina HiSeq 2000 and provides differential gene expression and pathway analysis. Powered by our unique bioinformatics capabilities, BGI’s RNA-Seq services deliver reliable data and analyses to our customers conducting drug response studies, biomarker detection, basic medical research, and drug R&D.
Benefits:
Compared to DNA microarray, BGI’s RNA-Seq (Quantification) service offers:
- Greater reproducibility
- Greater specificity - more accurate, no cross-hybridization, no restrictions to the available probes
- Greater sensitivity- detects 50% more genes, wider dynamic range.
- Strong bioinformatics analysis capabilities on RNA-Seq data
- Results compatible with common analysis software
BGI has validated the reproducibility, sensitivity, and detection range of RNA-Seq(Quantification).
Some results are as follows:
Materials: Human Brain Reference RNA (HBRR) from FirstChoice ®;
Universal Human Reference RNA (UHRR) from Stratagene
Standardized method of gene expression analysis: RPKM (Reads per kb per million reads) is used to quantify the gene expression level. The method is able to eliminate the influence of different gene lengths and sequencing discrepancy in the measurement.
High Reproducibility
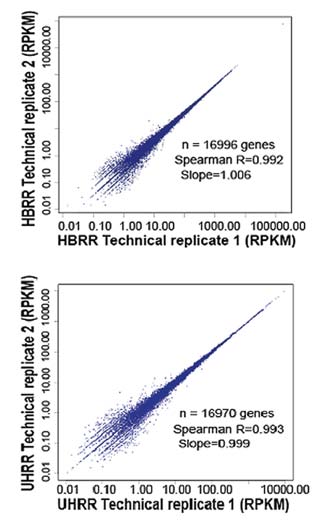
Figure 1: Technical Replicate Results of RNA-Seq (Quantification). From the technical replicate data, the spearman R is 0.992 and 0.993, respectively.
The results suggest that RNA-Seq has high reproducibility.
High Accuracy
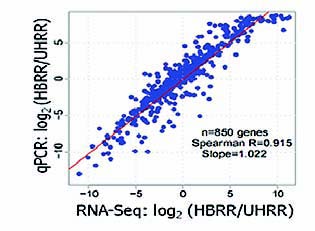
Figure 2: Correlation between results from RNA-Seq (Quantification) and qPCR RNA-Seq is a highly accurate quantitation method. The scatter-plot of figure 2 show a high correlation between results from RNA-Seq and qPCR. Spearman R is 0.915 and slope is 1.022.
High Sensitivity
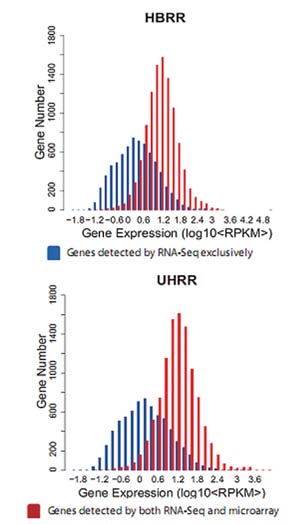
Figure 3: Detection Range of RNA-Seq (Quantification) is wider than microarray, and RNA-Seq can detect more low abundance genes than micriarray.
Bioinformatics:
Standard Bioinformatics Analysis
- Data filtering includes removing adaptors, contamination and low-quality reads from raw reads
- Assessment of sequencing
- Gene expression annotation.
Advanced Bioinformatics Analysis
- Differential gene expression analysis
- Expression pattern analysis of differentially expressed genes (DEGs)
- Gene ontology analysis of DEGs
- Pathway enrichment analysis of DEGs
- Protein-protein interaction network analysis
Custom Bioinformatics Analysis
- We can also perform customized analyses to meet the requirements of specific projects.
Sample Requirements:
- Sample condition: Integrated total RNA samples that have been treated with DNase; Avoid protein contamination during RNA isolation
- Sample quantity (for library construction): Total RNA ≥ 10 µg
- Sample concentration: ≥ 200 ng/µL
- Sample purity: OD260/280 = 1.8-2.2; OD260/230 ≥ 1.8; for animal RIN ≥ 7.0, for plant and fungi RIN ≥ 6.5; 28S:18S ≥ 1.0
Turnaround Time:
The standard turnaround time for the workflow (above) is 30 business days.

