PDX ToolKits
Cases
Technical Information
Contact Us / Wish List
 To better reflect human disease pathology in mouse models, patient-derived xenografts (PDX) have been widely used to evaluate new anti-cancer drugs for potential development in human clinical trials. One limitation of this approach is that the mouse genome is almost 90% homologous to the human genome, leading to possible contamination and complicating downstream bioinformatics analyses. The application of xenograft technology is further impeded by several technical issues, such as a lack of qualified paired controls for accurate variant profiling and a mixture of genetic factors from the host in the xenograft. To address these issues, BGI developed comprehensive patient-derived xenograft toolkit sets (“PDX Toolkits”) with a modular design comprised of tools encompassing all functions for HiSeq data from basic mapping to variant recalibration and annotation.
To better reflect human disease pathology in mouse models, patient-derived xenografts (PDX) have been widely used to evaluate new anti-cancer drugs for potential development in human clinical trials. One limitation of this approach is that the mouse genome is almost 90% homologous to the human genome, leading to possible contamination and complicating downstream bioinformatics analyses. The application of xenograft technology is further impeded by several technical issues, such as a lack of qualified paired controls for accurate variant profiling and a mixture of genetic factors from the host in the xenograft. To address these issues, BGI developed comprehensive patient-derived xenograft toolkit sets (“PDX Toolkits”) with a modular design comprised of tools encompassing all functions for HiSeq data from basic mapping to variant recalibration and annotation.
Benefits:
- Novel and efficient algorithm (PDXomics) to filter out mouse genome contaminants and acquire a highly accurate variant set of PDX models
- First-in-kind solution (PDXsnv) to identify germline mutations and predict somatic single nucleotide variations (SNVs) in the absence of normal tissues
- Comprehensive cancer genome database (18,406 human cancer samples sequenced at BGI) for cross-validation and auto-correction of genetic variants
- Robust bioinformatics pipeline to detect SNP, Indel, and CNV calling with high accuracy
- Integrative methods (four reliable bioinformatics tools available) to identify structural variations (SV)
- Cost-effective and rapid validation by incorporation of in situ and RNA-Seq validation into the pipeline
Figure 1. Key features of PDX Toolkits @ BGI
PDX Toolkits enable the prediction of somatic mutations without requiring the normal tissue controls, provide a more efficient method for eliminating mouse genome contamination, and enhance the validation of genetic variants using our comprehensive cancer genome databases.
1. Filter out mouse contaminants with PDXomics @ BGI
Based on our data from PDXomics@BGI, anywhere from 5%–33% of the sequencing reads from xenograft samples are actually contaminants from the mouse genome sequence. The amount of contamination varied between different models, different vendors, and in various cancer types. We found that there is an obvious concordance between DNA and RNA data.
Figure 2. Mouse Contamination Rates in Xenografts
2. Somatic SNV prediction in the absence of normal control tissue
Our PDXsnv algorithm is able to decrease the number of predicted somatic SNVs significantly from more than 3,500,000 (GATK results) to less than 20,000. Moreover, PDXsnv predicts the somatic SNVs of major cancer types with at least 75% sensitivity in the absence of corresponding normal controls, covers the known driver and suppressor genes, and detects novel SNVs.
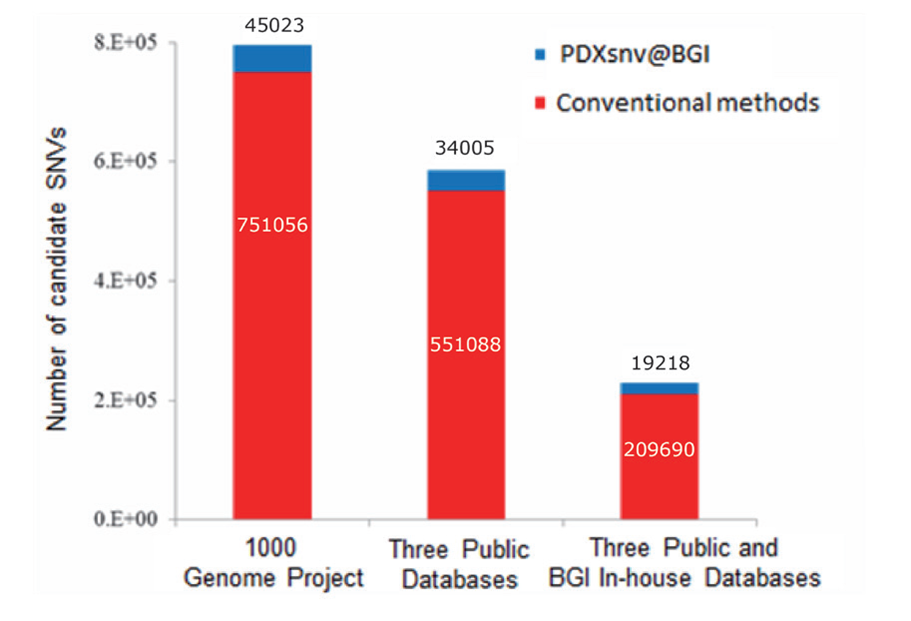
Figure 3. Efficient SNV Calling using PDXsnv @ BGI. 3 major public databases include latest dbSNP (v.137), 1000 Genome Project exp. validated mutations (2012), and ESP exomic database.
This figure shows that PDXsnv@BGI significantly reduces the number of candidate somatic SNVs:
- Our proprietary in-house database reduces the number to 209,690.
- Our unique machine learning algorithm further decreases the number to 19,218.
3. PDX genomic data is concordant with clinical samples
Somatic mutations of seven pairs of primary tumors and their corresponding xenograft samples are identified in the absence of normal control samples at an accuracy of >80% when analyzing a panel of cancer associated genes. PDXs are highly consistent with primary tumor samples in the variation patterns of cancer associated genes (Figure 4, upper panel). A more detailed analysis of one primary tumor-xenograft pair shows highly concordant gene expression (Figure 4, lower panel).
Figure 4. Comparisons between xenograft models and primary tumors show high similarities in patterns of genetic variations (upper panel) and gene expression profile (lower panel).
PDX Toolkit is a software package developed at BGI to reveal intrinsic mechanisms and features of PDX models systematically and comprehensively (Figure 5), which facilitates translational research and drug discovery.
Figure 5. Patient-derived Xenograft Toolkit sets (PDX Toolkits)
The toolkit offers a wide variety of tools (modules), including a basic mapping and removal of mouse contamination module, a statistics module at the sample level, a primary variant discovery and genotyping module, as well as powerful processing variant recalibrating and annotating modules (Table 1).
Table 1. Modules and their functions in PDX Toolkits
|
Module(s) |
Function(s) |
|
[Lane] |
Distinguish human reads from mouse reads with very high accuracy for downstream analysis using BGI’s self-developed PDXomics algorithms |
|
[MARSC] |
Merge files as defined by users Recalibrate variants with Gaussian error model |
|
[Queue] & [CNV] |
Identify SNPs, SNVs, Indels, and CNVs in PDX samples without requiring normal matched controls using BGI’s self-developed PDXsnv algorithms |
|
[SVcaller] |
Identify SVs from sequencing data using a combination of six methods of analysis (e.g., BreakDancerMax, Crest, Pindel) |
|
[SVir] |
Integrate all SV calling results from SVcaller and locate accurate breakpoints |
|
[AnnTools] |
Annotate variants from above modules and infer mechanisms for the involvement of SVs |


