- Genomics
- Transcriptomics
- Epigenomics
- Meta-omics
- Proteomics
- Single-Cell Sequencing
- Immune Repertoire Sequencing
- FFPE Samples

Immune Repertoire Sequencing
Cases
Technical Information
Contact Us / Wish List
 Immune repertoire refers to the sum total of functionally diverse B and T-cells in the circulatory system at any given moment (Wang et al. 2011). The diversity of immune repertoire is of vital importance for health. Immune Repertoire Sequencing (IR-SEQ) is to amplify the complimentary determining region (CDR) of B-cell receptor (BCR) or T-cell receptor (TCR) using multiple-PCR or 5’ RACE methods, followed by high-throughput sequencing. It is used to study the diversity of the immune system and the associations between immune repertoire and diseases.
Immune repertoire refers to the sum total of functionally diverse B and T-cells in the circulatory system at any given moment (Wang et al. 2011). The diversity of immune repertoire is of vital importance for health. Immune Repertoire Sequencing (IR-SEQ) is to amplify the complimentary determining region (CDR) of B-cell receptor (BCR) or T-cell receptor (TCR) using multiple-PCR or 5’ RACE methods, followed by high-throughput sequencing. It is used to study the diversity of the immune system and the associations between immune repertoire and diseases.
Benefits:
- Proprietary methods: we have designed specific primers for CDR3 V region and C region to amplify CDR3 region, and patents for TRA, IGH, and IGK/IGL have been applied successfully.
- High-fidelity amplification methods: multiple-PCR or 5’ RACE method is used to equivalently amplify different clones and the number of unique clones is as large as 105.
- Multiple sequencing platforms: Hiseq2000, Hiseq2500 and Roche 454 are available for different project requirements.
Applications:
- Vaccine development and efficacy
- Biomarker discovery
- Minimal residual disease detection
- Investigation of autoimmune diseases
- Transplant rejection and tolerance
References
1. C. Wang et al. J. Immunol. 186, 65.20 (2011)
Coming soon...
Workflow:
Peripheral blood mononucleated cells (PBMCs) are isolated from peripheral blood, followed by DNA or RNA extraction. Multiple-PCR or 5’RACE method is used to capture CDR3 region (5’ RACE can also capture CDR1 and CDR2), and then the captured region is sequenced on high-throughput platforms (Hiseq2000, Hiseq2500, Miseq or Roche 454).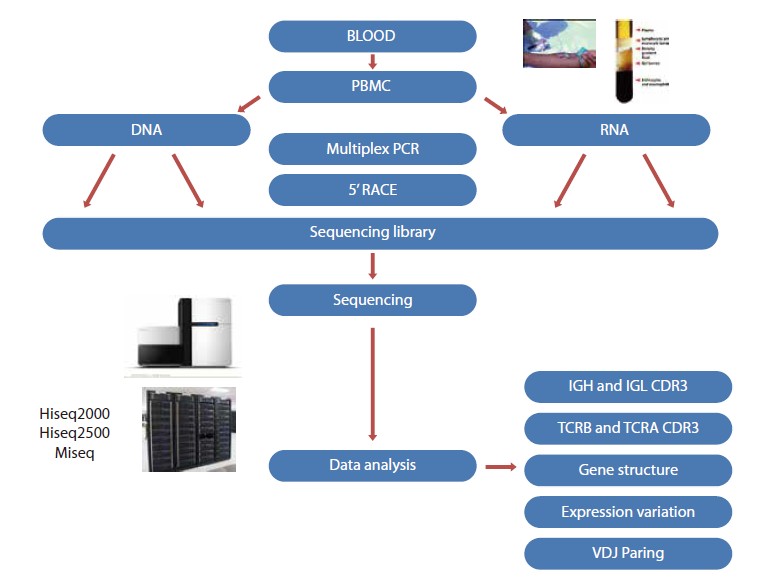
Bioinformatics Analysis:
Basic data processing
- Data filtering, removal of adapter contamination and low quality reads from raw reads
- Merge paired reads and remove background noise
- Output data statistics and evaluation of contents and quality of sequencing data
Alignment
- Alignment to V/D/J gene segments separately in IMGT database
- Realignment for best result
Structure analysis
- CDR sequence and base composition
- V/D/J recombination insertion and deletion
- Translate CDR sequences into peptides
Immune repertoire profiling
- IR expression profiling
- Diversity visualization, 2D and 3D histogram of V-J pairing
Expression differential analysis
- Differential analysis of diversity between samples (e.g. Simpson index, Shannon-Weaver index)
- Differential expression analysis of clones between samples (e.g. CDR3, V-D-J)
- Differential expression analysis of clones between groups (e.g. CDR3, V-D-J)
Customized bioinformatics analysis
We can perform other customized analyses to meet specific requirements.Sample Requirements:
Genomic DNA
- Purity: OD260/280=1.8~2.0, without degradation and RNA contamination
- Concentration: c ≥ 37.5ng/μL
- Quantity: 2.5 μg ≤ amout < 5 μg (for library construction once); amount ≥ 5 μg (for library construction more than twice)
RNA sample
- Purity: RIN ≥ 7.0, 28S/18S ≥ 1.0, the baseline is smooth and 5S peak is normal
- Concentration: c ≥ 200ng/μL
- Quantity: 5 μg ≤ amount

