- Genomics
- Transcriptomics
- Epigenomics
- Meta-omics
- Proteomics
- Single-Cell Sequencing
- Immune Repertoire Sequencing
- FFPE Samples

Single-Cell RNA Sequencing
Cases
Technical Information
Contact Us / Wish List
 Scientific research through traditional transcriptome analysis of early-stage embryos, stem cells, cancers, immune cells, and developing neurons is limited, because the amount of sample that is obtained does not meet the minimum requirements for traditional NGS, which is probably due to extremely little or immensely high heterogeneity. Thus, the need for single-cell transcriptome analysis is particularly urgent. Single-cell RNA-Seq quantifies the mRNA of a single cell through single-tube reverse transcription and PCR amplification, allowing several micrograms of cDNA to be collected for traditional library construction and Hiseq2000 sequencing. Single-cell RNA-Seq is a novel application of RNA-Seq that is capable of sequencing a single cell or infinitesimal amounts of sample.
Scientific research through traditional transcriptome analysis of early-stage embryos, stem cells, cancers, immune cells, and developing neurons is limited, because the amount of sample that is obtained does not meet the minimum requirements for traditional NGS, which is probably due to extremely little or immensely high heterogeneity. Thus, the need for single-cell transcriptome analysis is particularly urgent. Single-cell RNA-Seq quantifies the mRNA of a single cell through single-tube reverse transcription and PCR amplification, allowing several micrograms of cDNA to be collected for traditional library construction and Hiseq2000 sequencing. Single-cell RNA-Seq is a novel application of RNA-Seq that is capable of sequencing a single cell or infinitesimal amounts of sample.
Benefits:
- Accommodates an infinitesimal amount of sample input, which is not possible with traditional sequencing protocols.
- Single-tube amplification greatly reduces sample loss. This benefit allows a minimum amount of sample to be required, ensures high sensitivity for library construction, avoids the loss of transcripts that are expressed with low abundance, and reduces data bias.
- Provides reliable data for subsequent analysis.
- More than 90% of genes can be detected.
- A greater number of low-abundance transcripts can be detected than using microarray.
- The technique is highly repeatable.
BGI has validated the sensitivity, reproducibility, and detection range of single-cell RNA-Seq.
High Sensitivity (number of detected genes)
Comparison of the single-cell RNA-Seq with a traditional cDNA microarray showed that RNA-Seq could detect a much greater number of genes than that detected by the cDNA microarray. The blue circle represents those genes detected by the cDNA microarray, while the red circle represents those genes detected by single-cell RNA-Seq.
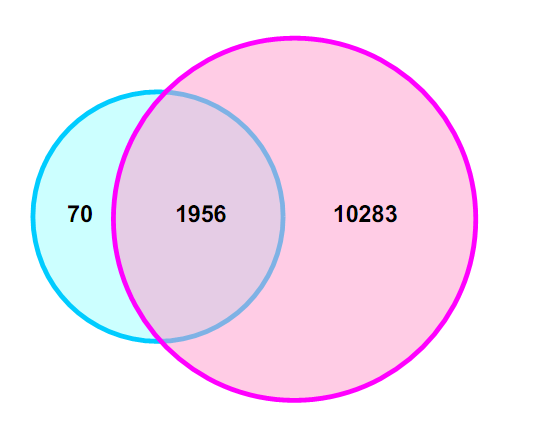
Fig 1. Comparison of number of detected genes between single-cell RNA-Seq and microarray in one oocyte
High Reproducibility
The consistency of gene expression (Pearson correlation coefficient) between adjacent blastomeres from the same zygote was greater than 80%.
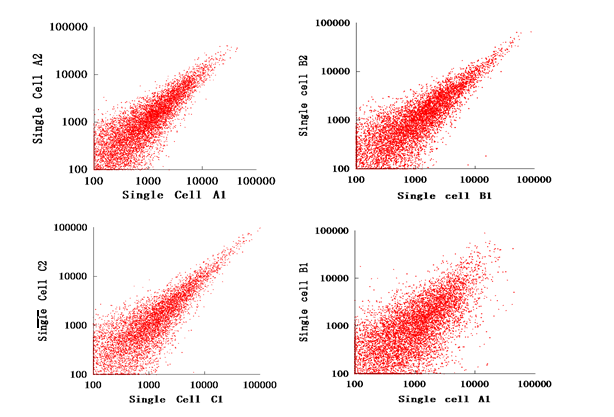
Fig 2. The correlation between different single cells. Every plot stands for the transcript mapped by at least 100 reads. (A-C)The Pearson Correlation Coefficient of cells from the same embryo is greater than 0.8. (D) By contrast, the Pearson Correlation Coefficient between cell B1 and cell A1 is only 0.6509.
High Throughput
Greater than 90% of genes can be detected with 30 million (M) raw reads. The abscissa shows the number of reads; the ordinate shows the percentage of genes that were detected. The two lines in blue and red stand for saturation curve of two different samples. So we suggest 30M reads per sample for single cell RNA-seq.
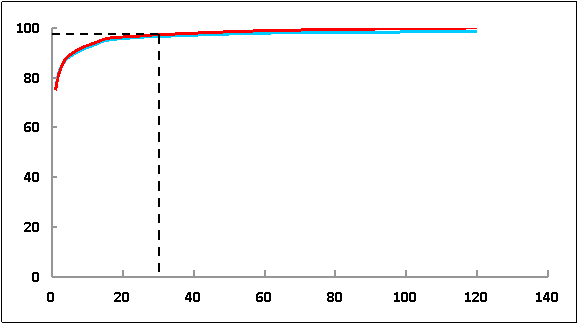
Fig 3. Saturation analysis of sequencing
Pipeline of single-cell RNA-seq
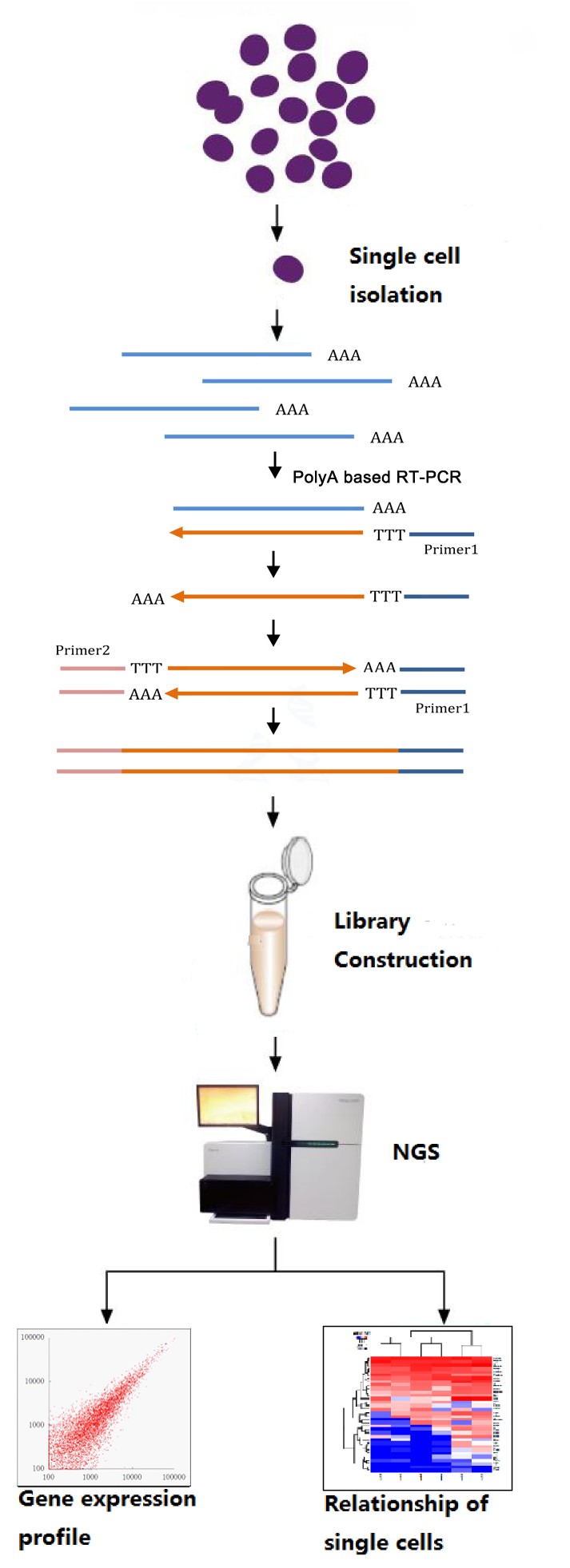
Bioinformatics Analysis:
Standard Bioinformatics Analysis
- Data filtering includes removing adaptors, contamination, and low-quality reads from raw reads
- Assessment of sequencing (statistics of raw reads, sequencing saturation analysis, and analysis of the distribution of reads for a reference gene)
- Gene expression annotation (gene coverage, sequencing depth, etc.)
- Differential gene expression analysis (two or more samples should be provided)
- Expression pattern analysis of differentially expressed genes (DEGs)
- Gene ontology enrichment analysis of DEGs
- Pathway enrichment analysis of DEGs
Customized Bioinformatics Analysis
We can perform other customized analyses to meet the requirements of specific projects.
Sample Requirements:
Singe-cell sample
| Level A | Mammalian oocyte, single blastomere from one-cell stage to eight-cell stage (e.g. single cell of human/mouse early-stage embryo) | Has been tested, success rate > 90% |
| Level B | Single mammalian cells from morula to blastula; both free single cells and cell lines with diameters greater than 10 μm (e.g. circulating tumor cells) | No test has been performed; theoretically executable with relatively low risk |
| Level C | Normal tissue cells | No test has been performed; lacks theoretical support, therefore high risk |
| Level D | Special tissues/structural cells, e.g. cells from vegetative pole of ovipara |
Has been tested, impracticable |
RNA sample
- Sample condition:Integrated total RNA samples (no mRNA isolation). Avoid protein contamination during RNA isolation.
- Sample quantity (for library construction once): total RNA ≥ 10 pg
- Sample concentration: ≥ 10 pg/μL
Turnaround Time:
The average turnaround time for RNA-Seq on 50 cells is about 40 workdays.
Completion Indicator:
Completion is indicated by the number of clean reads. The goals are individualized for each project.

